[ICLR 24] AnomalyCLIP: Object-agnostic Prompt Learning for Zero-shot Anomaly Detection
by Qihang Zhou*, Guansong Pang*, Yu Tian, Shibo He, Jiming Chen.
03.19.2024: Code has been released !!!
08.08.2024: Update the code for testing one image.
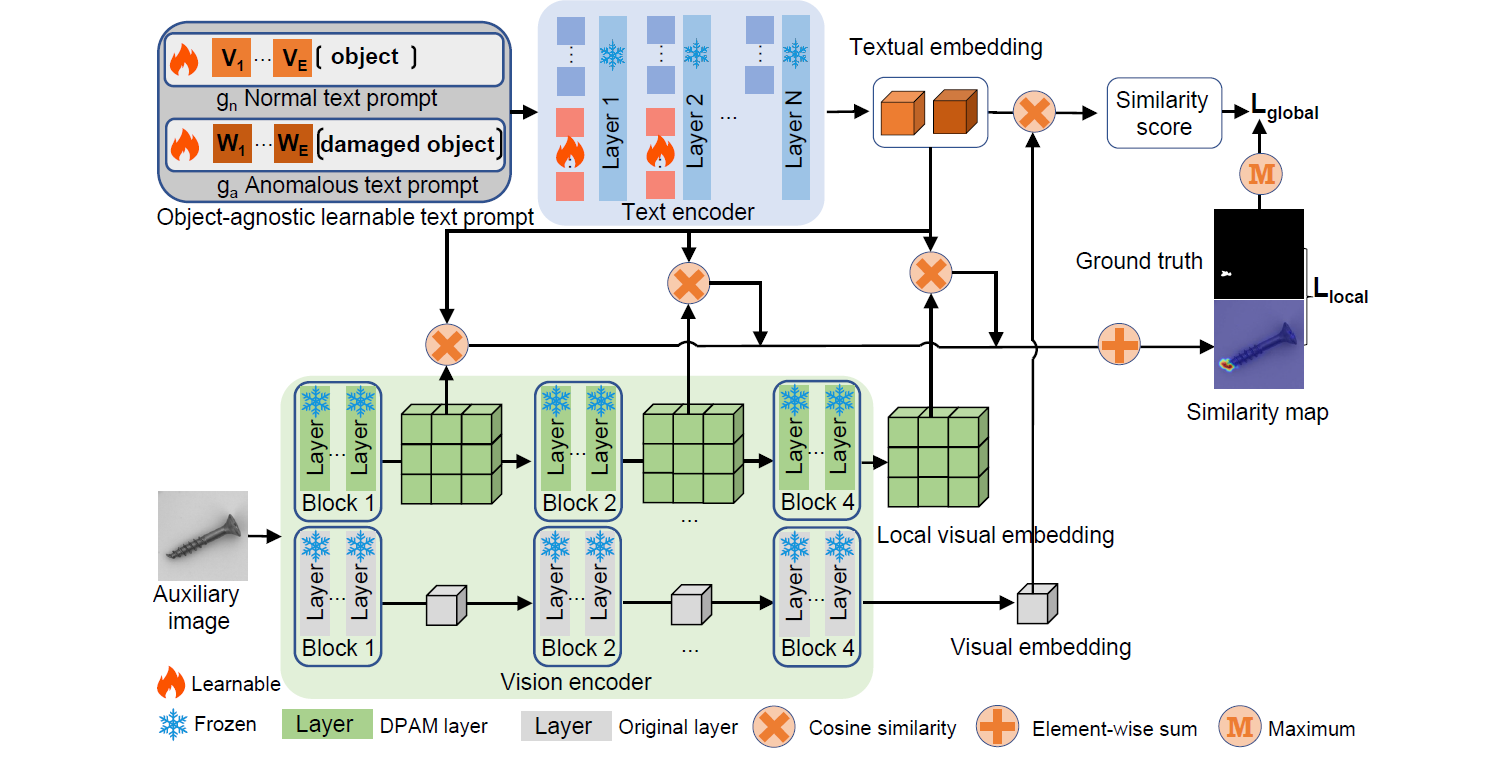
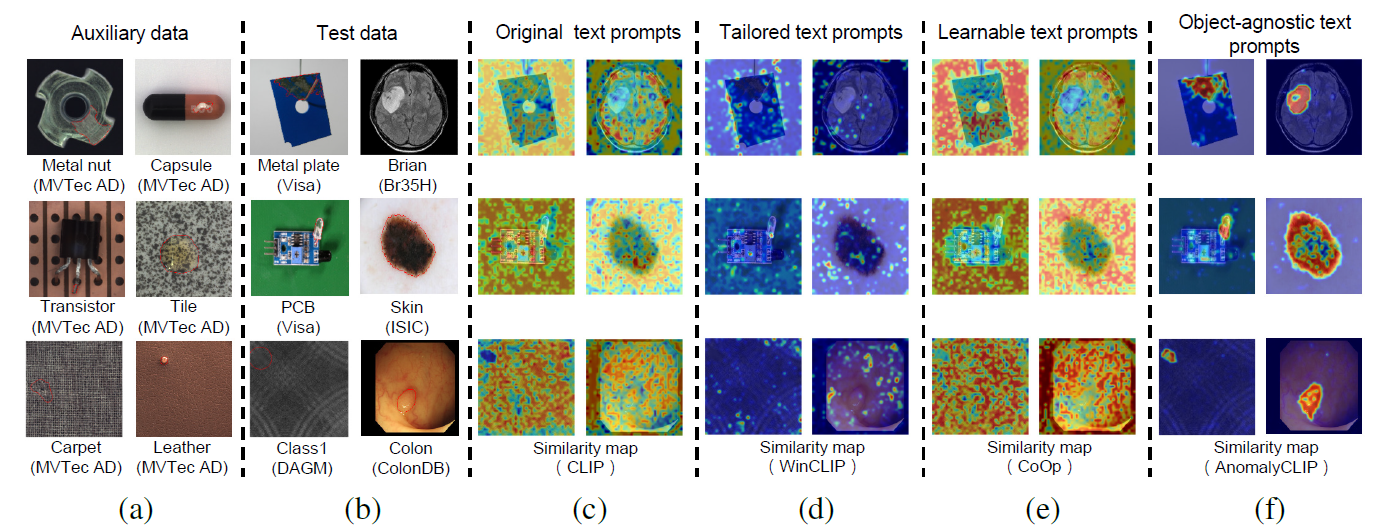
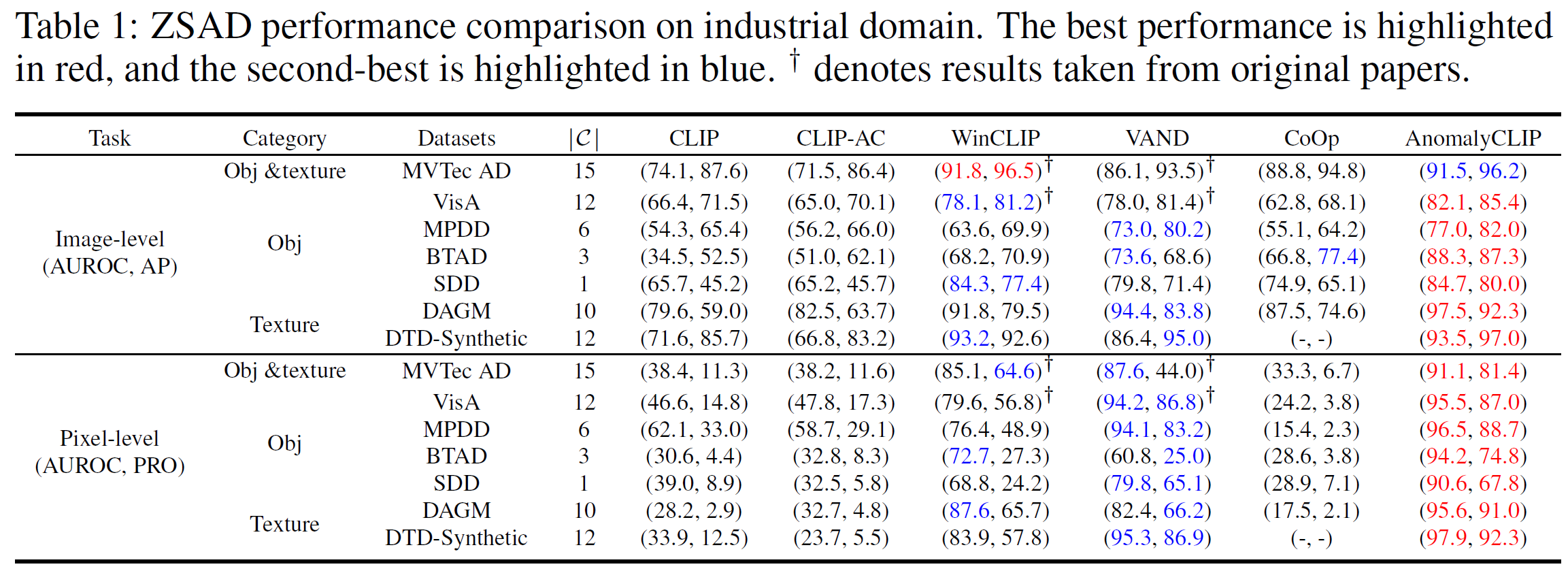
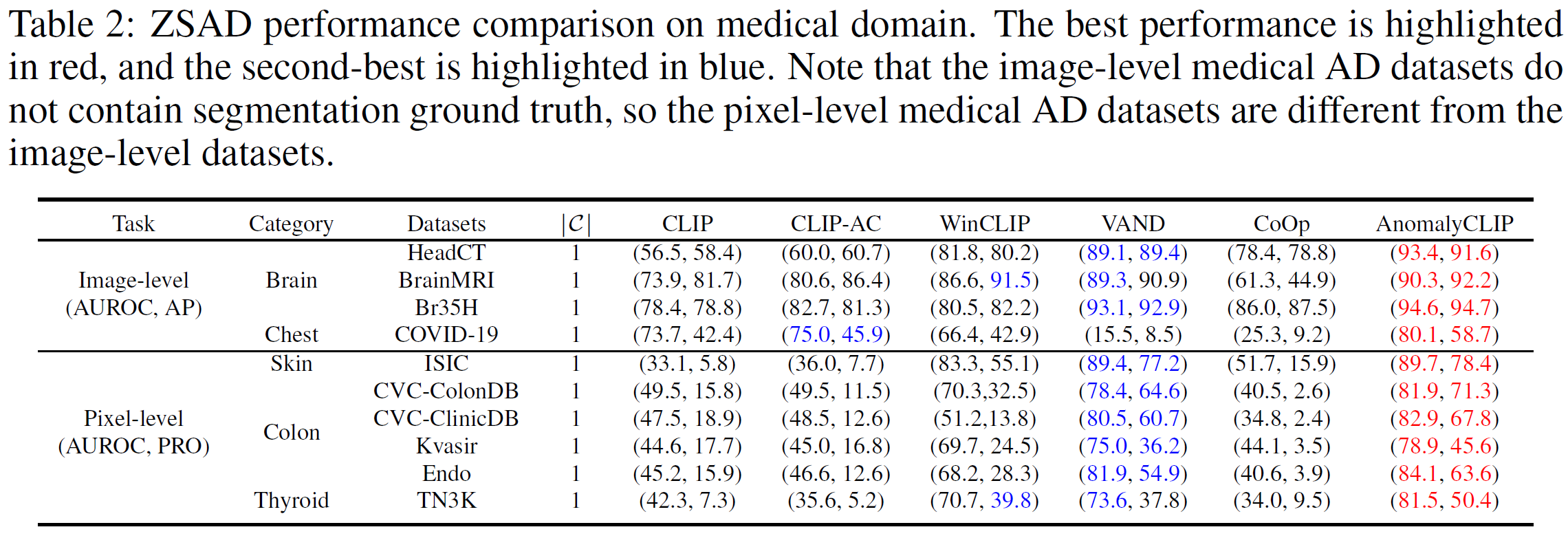
Zero-shot anomaly detection (ZSAD) requires detection models trained using auxiliary data to detect anomalies without any training sample in a target dataset. It is a crucial task when training data is not accessible due to various concerns, e.g., data privacy, yet it is challenging since the models need to generalize to anomalies across different domains where the appearance of foreground objects, abnormal regions, and background features, such as defects/tumors on different products/organs, can vary significantly. Recently large pre-trained vision-language models (VLMs), such as CLIP, have demonstrated strong zero-shot recognition ability in various vision tasks, including anomaly detection. However, their ZSAD performance is weak since the VLMs focus more on modeling the class semantics of the foreground objects rather than the abnormality/normality in the images. In this paper we introduce a novel approach, namely AnomalyCLIP, to adapt CLIP for accurate ZSAD across different domains. The key insight of AnomalyCLIP is to learn object-agnostic text prompts that capture generic normality and abnormality in an image regardless of its foreground objects. This allows our model to focus on the abnormal image regions rather than the object semantics, enabling generalized normality and abnormality recognition on diverse types of objects. Large-scale experiments on 17 real-world anomaly detection datasets show that AnomalyCLIP achieves superior zero-shot performance of detecting and segmenting anomalies in datasets of highly diverse class semantics from various defect inspection and medical imaging domains. All experiments are conducted in PyTorch-2.0.0 with a single NVIDIA RTX 3090 24GB.
Download the dataset below:
Industrial Domain: MVTec, VisA, MPDD, BTAD, SDD, DAGM, DTD-Synthetic
Medical Domain: HeadCT, BrainMRI, Br35H, COVID-19, ISIC, CVC-ColonDB, CVC-ClinicDB, Kvasir, Endo, TN3K.
Google Drive link (frequently requested dataset): SDD, Br35H, COVID-19
Take MVTec AD for example (With multiple anomaly categories)
Structure of MVTec Folder:
mvtec/ │ ├── meta.json │ ├── bottle/ │ ├── ground_truth/ │ │ ├── broken_large/ │ │ │ └── 000_mask.png | | | └── ... │ │ └── ... │ └── test/ │ ├── broken_large/ │ │ └── 000.png | | └── ... │ └── ... │ └── ...
cd generate_dataset_json python mvtec.py
Take SDD for example (With single anomaly category)
Structure of SDD Folder:
SDD/ │ ├── electrical_commutators/ │ └── test/ │ ├── defect/ │ │ └── kos01_Part5_0.png | | └── ... │ └── good/ │ └── kos01_Part0_0.png │ └── ... │ └── meta.json
cd generate_dataset_json python SDD.py
Select the corresponding script and run it (we provide all scripts for datasets that AnomalyCLIP reported). The generated JSON stores all the information that AnomalyCLIP needs.
Create a new JSON script in fold generate_dataset_json according to the fold structure of your own datasets.
Add the related info of your dataset (i.e., dataset name and class names) in script dataset.py
Quick start (use the pre-trained weights)
bash test.sh
Train your own weights
bash train.sh
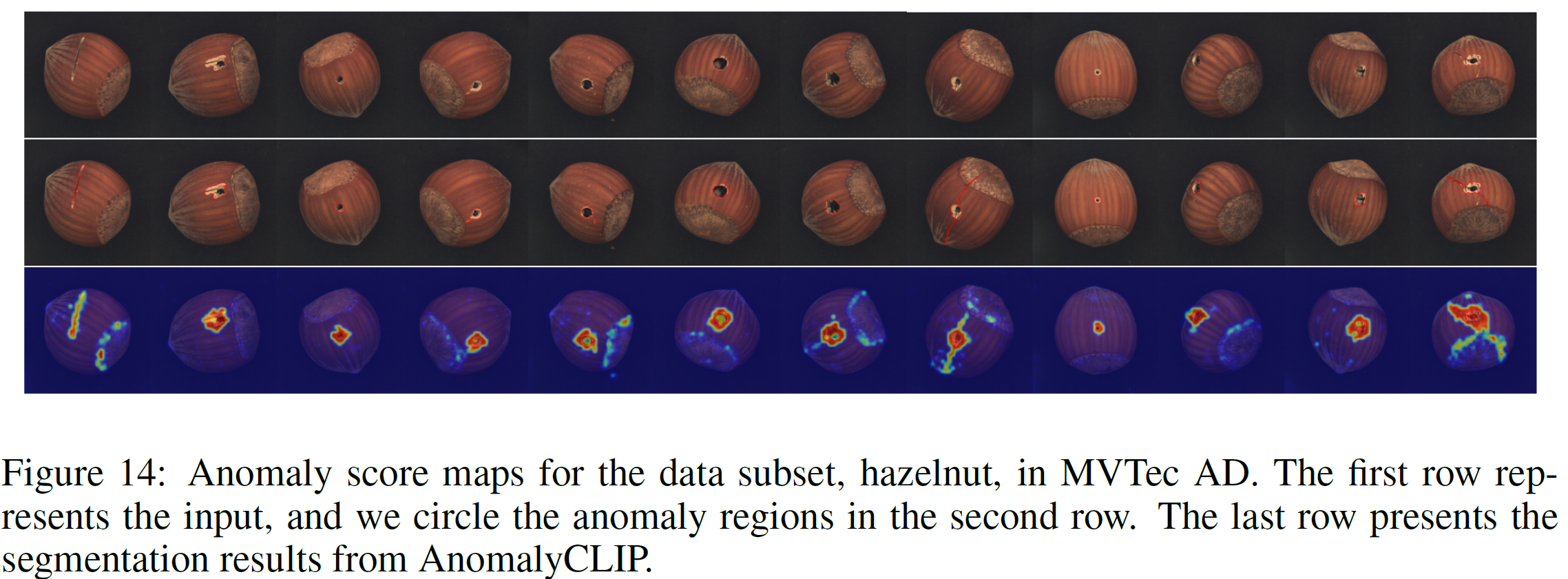

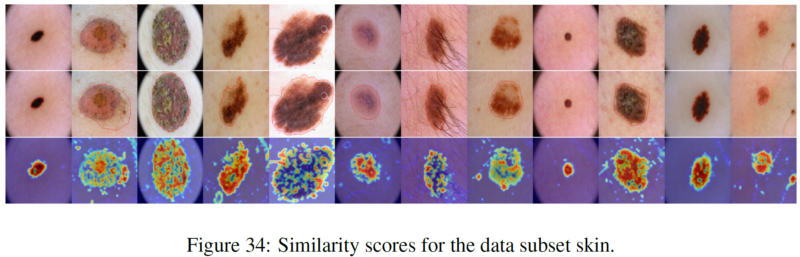
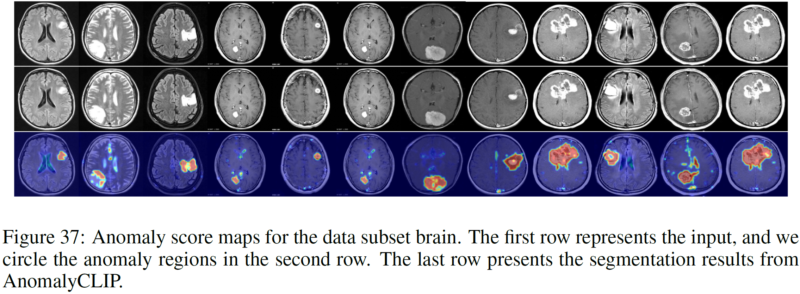
We thank for the code repository: open_clip, DualCoOp, CLIP_Surgery, and VAND.
If you find this paper and repository useful, please cite our paper.
@inproceedings{zhou2023anomalyclip,
title={AnomalyCLIP: Object-agnostic Prompt Learning for Zero-shot Anomaly Detection},
author={Zhou, Qihang and Pang, Guansong and Tian, Yu and He, Shibo and Chen, Jiming},
booktitle={The Twelfth International Conference on Learning Representations},
year={2023}
}