Last updated: 9/30/2024
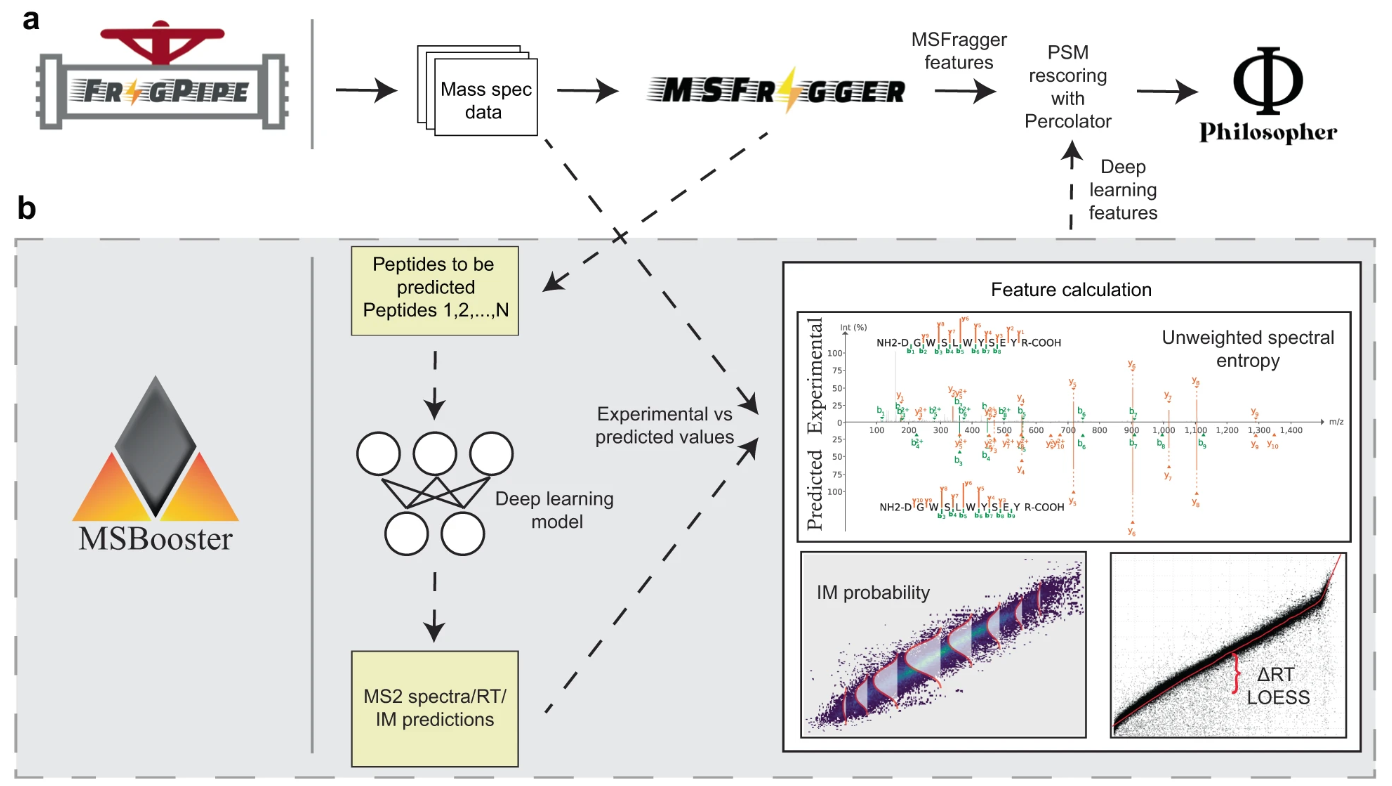
MSBooster is a tool for incorporating spectral libary predictions into peptide-spectrum match (PSM) rescoring in bottom-up tandem liquid chromatography mass spectrometry proteomics data. It is roughly broken into 4 steps:
Peptide extraction from PSMs in search results, and formatting for machine/deep learning (ML/DL) predictors' input files
Calling the prediction model(s) and saving the output
Feature calculation
Addition of new features to the search results file
MSBooster is compatible with many types of database searches, including HLA immunopeptidomics, DDA and DIA, and single cell proteomics. It is incorporated into FragPipe and is included in many of its workflows. MSBooster was developed with other FragPipe tools in mind, such as FragPipe-PDV.
MSBooster is equipped to handle multiple input file formats and models:
| Mass spectrometer output |
|---|
| .mzML |
| .mgf |
| PSM file |
|---|
| .pin |
| .pepXML (in progress) |
| Prediction model |
|---|
| DIA-NN |
| Koina models |
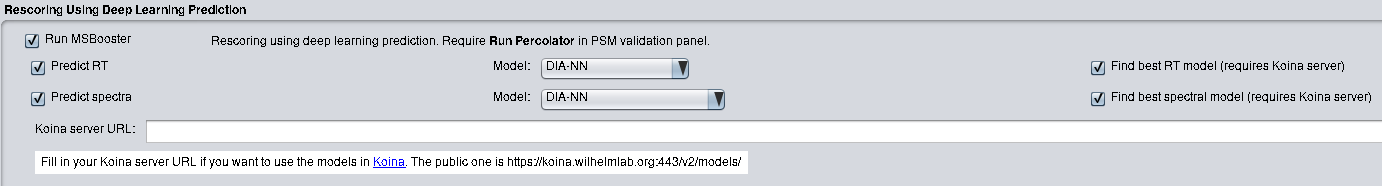
MSBooster can be run in Windows and Linux systems. If using FragPipe, no other installation steps are needed besides installing FragPipe. MSBooster is located in the "Validation" tab. Choose to enable retention time features with "Predict RT" and MS/MS spectral features with "Predict spectra". Please refer to the FragPipe documentation for how to run an analysis.
If using standalone MSBooster to run in the command line, please download the latest jar file from Releases. MSBooster also requires DIA-NN for MS/MS and RT prediction. Please install DIA-NN and take note of the path to the DIA-NN executable (ex. DiaNN.exe for Windows, diann-1.8.1.8 for Linux).
You can run MSBooster using a command similar to the following:
java -jar MSBooster-1.2.1.jar --paramsList msbooster_params.txt
The minimum parameters needing to be passed are:
- DiaNN (String): path to DIA-NN executable (if using DIA-NN model, which is the MSBooster default) - mzmlDirectory (String): path to mzML/mgf files. Accepts multiple space-separated folder and files - pinPepXMLDirectory (String): path to pin files. Accepts multiple space-separated folder and files. If using in FragPipe, place the pin and pepXML files in the same folder
While you can individually pass these parameters, it is easier to place one on each line of the paramsList file. Please refer to msbooster_params.txt for a template.
The parameters below are for general use. Koina-specific parameters are in the Koina documentation
paramsList (String): location to text file containing parameters for this run
fragger (String): file path of fragger.params file from the MSFragger run. MSBooster will read in multiple parameters
and adjust internal parameters based on them, such as fragment mass error tolerance and mass offsets
outputDirectory (String): where to output the new files
editedPin (String): MSBooster will name the new file based on the ones provided. For example, A.pin will have a counterpart
called A_edited.pin. To change from the default of "edited", provide a new string here
renamePin (int): whether to generate a new pin file or rewrite the old one. Default here is 1, which will not overwrite.
Setting this to 0 will overwrite the old pin file
deletePreds (boolean): whether to delete the files storing model predictions after finishing a succesful run. By default, set
to false. Set to true if you wish to delete these
loadingPercent (int): how often to report progress on tasks using a progress reporter. By default, set to 10, meaning an
update will be printed every 10%.
numThreads (int): number of threads to use. By default set to 0, which uses all available threads minus 1
splitPredInputFile (int): only used when DIA-NN predictions fail due to an out of memory error (137). By default, set
to 1, but you can increase this to specify how many smaller files the DIA-NN input file should be broken up into. Each
file will then be predicted sequentially, easy the memory burden
plotExtension (String): what file format plots should be in. png by default, and pdf is also allowed
features (String): list of features to be calculated. Case-sensitive, comm-separated without spaces in between.
Default is "predRTrealUnits,unweightedSpectralEntropy,deltaRTLOESS"
spectraPredFile (String): if you are reusing old spectral predictions (e.g. from DIA-NN or Koina), you can specify the file
location here
RTPredFile (String): same as spectraPredFile, but for RT predictions
IMPredFile (String): same as spectraPredFile, but for IM predictions
spectraModel (String): which spectral prediction model to use
rtModel (String): same as spectraModel, but for RT
imModel (String): same as spectraModel, but for IM
useSpectra (boolean): whether to use spectral prediction-based features. Set to true by default
useRT (boolean): whether to use RT prediction-based features. Set to true by default
useIM (boolean): whether to use IM prediction-based features. Set to false by default
ppmTolerance (float): fragment error ppm tolerance (default 20ppm)
matchWithDaltons (boolean): whether to match predicted and observed fragments in Daltons (default false)
DaTolerance (float): how many daltons around the predicted peak to look for experimental peak (default 0.05)
useTopFragments (boolean): whether to filter spectral prediction to the N highest intensity peaks (default true)
topFragments (int): up to how many predicted fragments should be used for feature calculation (default 20). Only
applied if useTopFragments is true
removeRankPeaks (boolean): Set to true by default, which filters out fragments from the experimental spectra once
matched. If false, experimental fragments can be matched by multiple PSMs from the same scan
useBasePeak (boolean): whether a lower limit should be applied to MS2 predictions to only use fragments with higher
intensity (default true)
percentBasePeak (float): percent at which fragment with intensity of some percent of base peak intensity is included
in similarity calculation. Only applied if useBasePeak is true (default 1)
loessEscoreCutoff (float): expectation value cutoff used for first pass at collecting PSMs for RT/IM calibration.
Default is 10^-3.5, or approximately 0.000316
rtLoessRegressionSize (int): maximum number of PSMs used for RT LOESS calibration (default 5000)
imLoessRegressionSize (int): same as rtLoessRegressionSize but for IM (default 1000)
minLoessRegressionSize (int): minimum number of PSMs needed to attempt LOESS RT/IM calibration (default 100). If fewer than
this number of PSMs are available, linear regression is used instead
minLinearRegressionSize (int): minimum number of PSMs needed to attempt linear regression RT/IM calibration (default 10).
If fewer than this number of PSMs are available, no calibration is attempted
loessBandwidth (String): list of bandwidths to try for RT/IM LOESS calibration (default 0.01,0.05,0.1,0.2). This must
be comma-separated with no spaces in between
regressionSplits (int): number of cross validations used for RT/IM LOESS calibration (default 5)
massesForLoessCalibration (String): masses for mass shifts that should be fit to their own calibration curves. List
is comma-separated with no spaces in between. The masses should be written to the same number of digits as in the PIN file
loessScatterOpacity (float): opacity of scatter plots in LOESS calibration figures, from 0 to 1 (default 0.35)
.pin file with new features. By default, new pin files will be produced ending in "_edited.pin". The default features used are "unweighted_spectral_entropy", "delta_RT_loess", and "pred_RT_real_units". If ion mobility features are enabled, "delta_IM_loess" and "ion_mobility" will also be included
spectraRT.tsv and spectraRT_full.tsv: input files for DIA-NN prediction model
spectraRT.predicted.bin: a binary file with predictions from DIA-NN to be used by MSBooster for feature calculation. If using FragPipe-PDV, these files are used to generate mirror plots of experimental and predicted spectra
MSBooster produces multiple graphs that can be used to further examine how your data compares to model predictions.
MSBooster_plots folder:
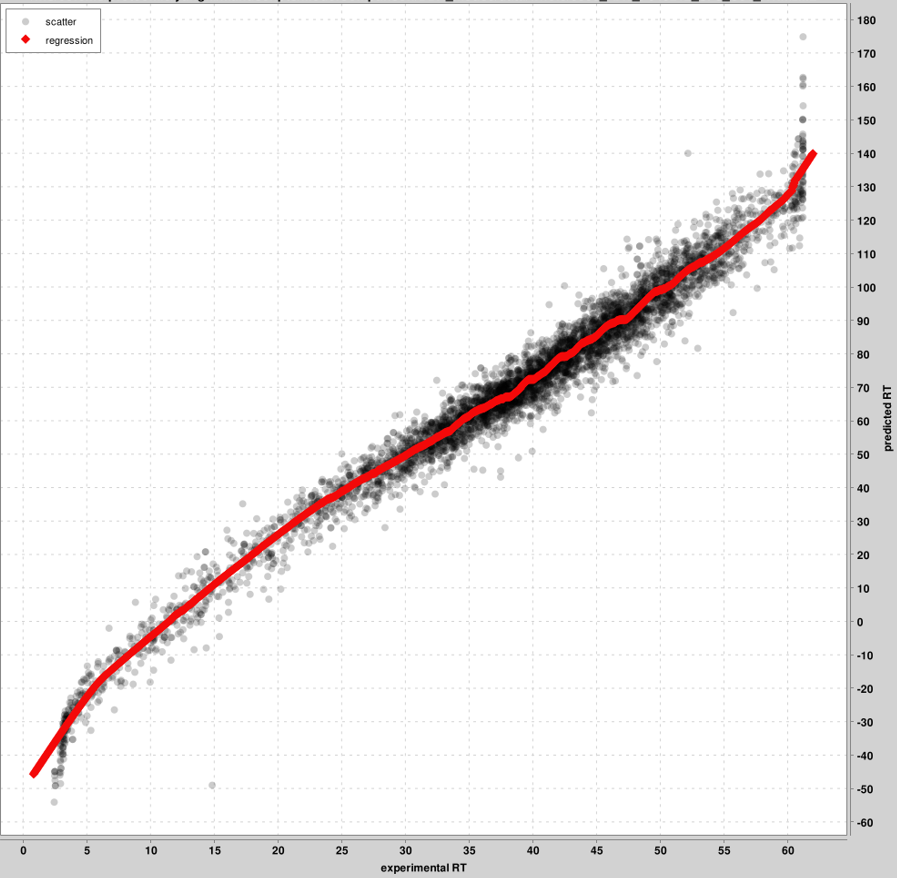
RT_calibration_curves: up to the top 5000 PSMs will be used for calibration between the experimental and predicted RT scales. These top PSMs are presented in the graph, not all PSMs. One graph will be produced per pin file
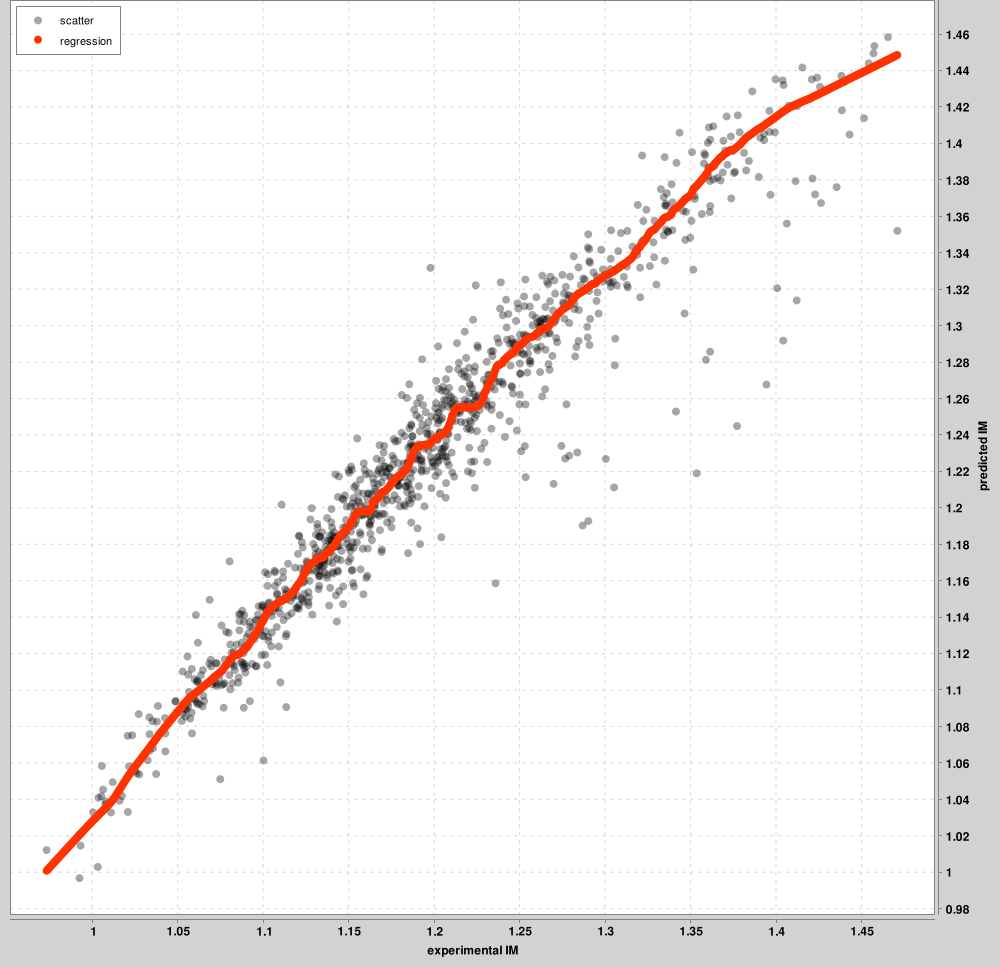
IM_calibration_curves: up to the top 1000 PSMs will be used for calibration between the experimental and predicted IM scales. These top PSMs are presented in the graph, not all PSMs. A separate curve will be learned for each charge state. The figure below is an example for charge 2 precursors
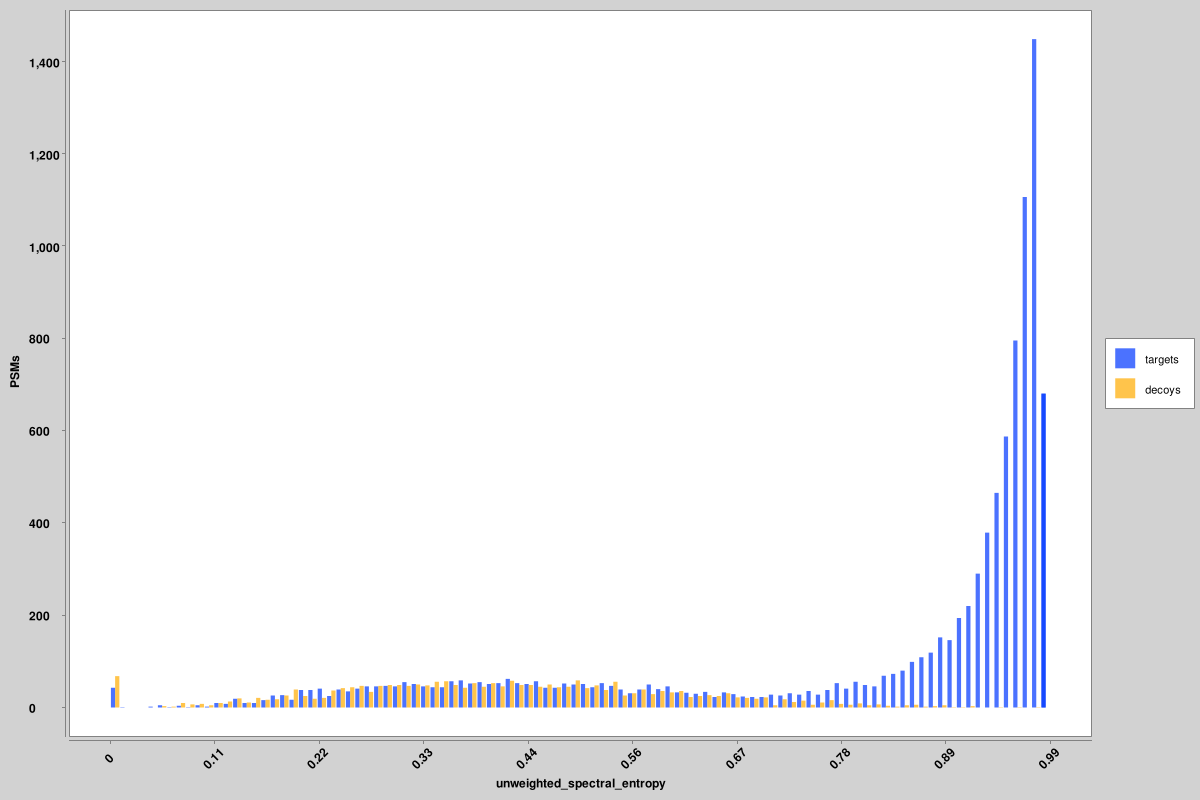
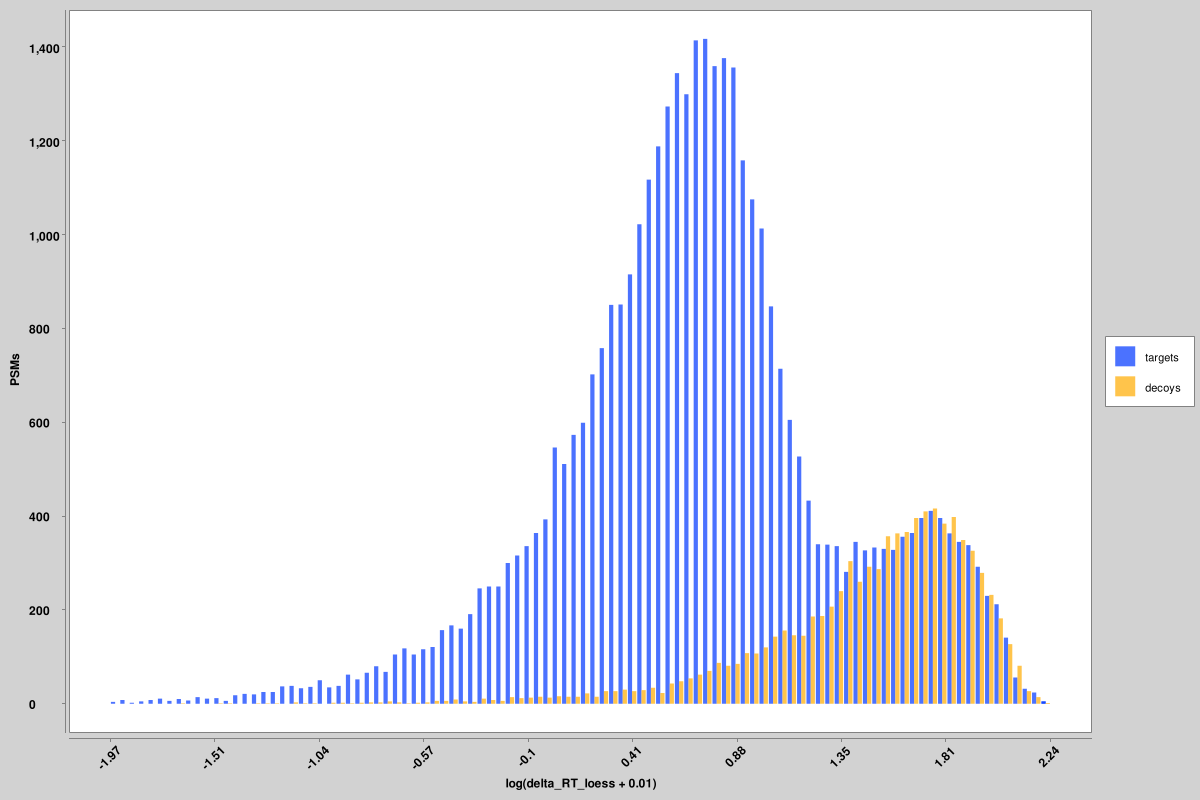
score_histograms: overlayed histograms of all target and decoy PSMs for each pin file. Some features are plotted here on a log scale for better visualization of the bimodal distribution of true and false positives, but the original value is what is used in the pin files, not the log-scaled version. Shown here are histograms for the unweighted spectral entropy and delta RT scores, but similar ones are produced for all features
Use peptide prediction models from Koina for MSBooster feature generation: https://fragpipe.nesvilab.org/docs/tutorial_koina.html
Reading in predictions from any model via MGF files
Documentation on all allowed features and how to QC them with graphical output
Please cite the following when using MSBooster: https://www.nature.com/articles/s41467-023-40129-9