The implementation of DiffAbXL benchmarked in the paper: Exploring Log-Likelihood Scores for Ranking Antibody Sequence Designs.
| Rank | Models | Absci HER2 | Nature | AZ Target-2 | Ave. ? | ||
|---|---|---|---|---|---|---|---|
| Zero Shot | SPR Control | HEL | HER2 | ||||
| 1 | DiffAbXL-A-DN | 0.43 | 0.22 | 0.62 | 0.37 | 0.41 | 0.41 |
| 2 | DiffAbXL-A-SG | 0.46 | 0.22 | 0.64 | -0.38 | 0.43 | 0.274 |
| 3 | DiffAbXL-H3-DN | 0.49 | 0 | 0.52 | -0.08 | 0.37 | 0.26 |
| 4 | IgBlend (struct. only) | 0.40 | 0.21 | 0.54 | -0.30 | 0.31 | 0.232 |
| 5 | Antifold | 0.43 | 0.22 | 0.4 | -0.47 | 0.38 | 0.192 |
| 6 | DiffAbXL-H3-SG | 0.48 | 0 | 0.4 | -0.41 | 0.29 | 0.152 |
| 7 | ESM | 0.29 | 0 | 0 | 0.18 | 0.27 | 0.148 |
| 8 | DiffAb | 0.34 | 0.21 | 0 | -0.14 | 0.22 | 0.126 |
| 9 | AbLang2 | 0.3 | 0 | 0 | -0.07 | 0.36 | 0.118 |
| 10 | IgBlend (seq. only) | 0.27 | 0 | 0 | -0.1 | 0.36 | 0.106 |
| 11 | AbLang | 0.3 | 0 | 0 | -0.13 | 0.35 | 0.104 |
| 12 | dyMEAN | 0.37 | 0.15 | 0 | 0 | 0 | 0.104 |
| 13 | AbX | 0.28 | 0.19 | 0 | 0 | 0 | 0.094 |
| 14 | AntiBERTy | 0.26 | 0 | 0 | -0.17 | 0.35 | 0.088 |
| 15 | MEAN | 0.36 | 0 | 0 | 0.02 | 0 | 0.076 |
| 16 | ESM-IF | 0 | -0.27 | 0 | -0.53 | 0.42 | -0.076 |
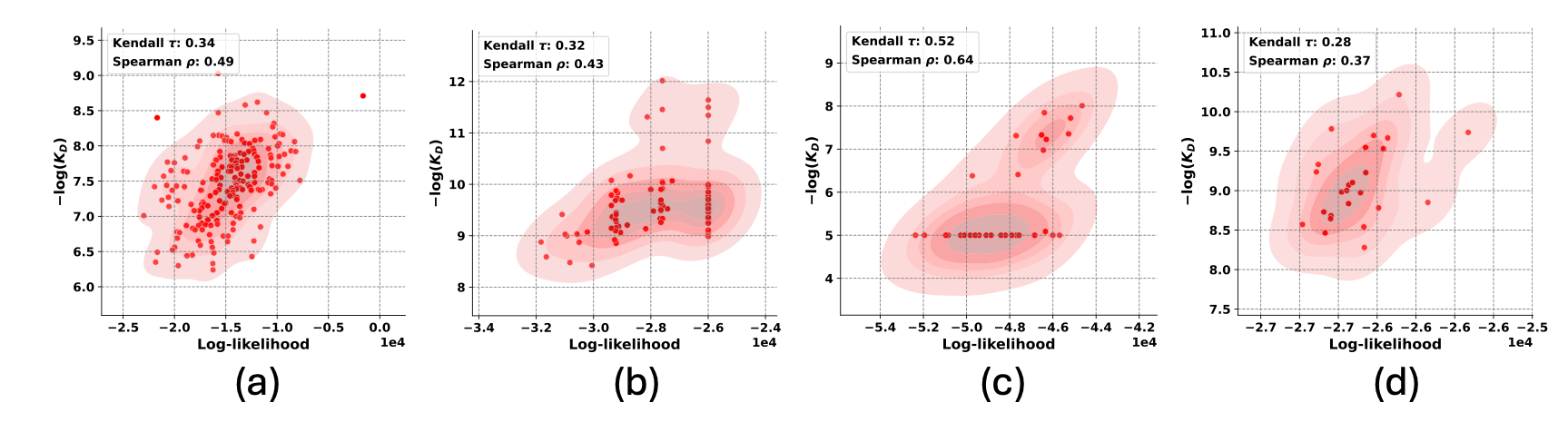
Figure-1: Results for DiffAbXL: a) DiffAbXL-H3-DN for Absci zero-shot HER2 data b) DiffAbXL-A-SG for AZ Target-2, c) DiffAbXL-A-SG for Nature HEL, d) DiffAbXL-A-DN for Nature HER2.
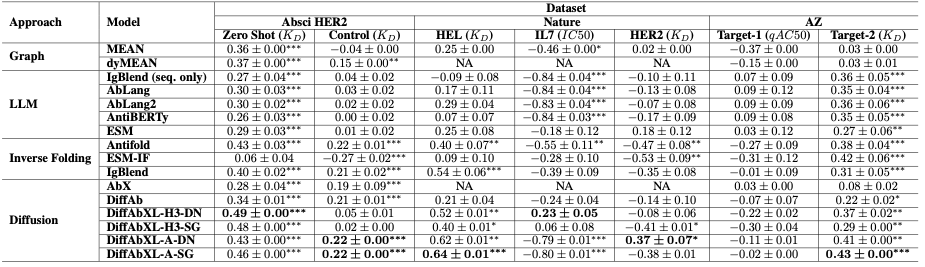
Table-1: Summary of the results for Spearman correlation. Abbreviations: DN: De Novo mode, SG: Structure Guidance mode, NA: Epitope or complex structure required, but not available. *, **, *** indicate p-values under 0.05, 0.01 and 1e-4 respectively.
To make it easier for us to benchmark your model, we recommend that you implement an interface as a Python method in a class that we can easily integrate with our evaluation pipeline. The method should accept the following inputs:
The method should return a dictionary containing:
Here's a basic template in Python for implementing this interface:
def benchmark(self, sequences, structure=None, mask=None, **kwargs):
"""
Benchmark the model on provided antibody sequences and structures.
Parameters:
sequences (list of str): List of antibody sequences.
structure (optional): Path to a PDB file. Currently, only one PDB file is provided per target dataset.
The PDB file may contain either just the antibody or an antibody-antigen complex,
depending on the dataset.
mask (optional): Binary list or array indicating the regions of interest in the sequences for metric calculations.
kwargs (optional): Additional parameters required by the model.
Returns:
dict: A dictionary containing log-likelihood scores and other relevant metrics.
"""
passPlease make sure that your model outputs the log-likelihood scores in a format we can use directly for benchmarking antibody sequence designs. This will help us compare your model's performance across our datasets efficiently.
There is one configuration file: sabdab.yaml, which can be used to change any of the parameters. You can train the model by using:
python train.py # For training.
- train.py
- src
|-model.py
- config
|-sabdab.yaml
- utils
|-load_data.py
|-arguments.py
|-model_utils.py
|-loss_functions.py
...
- data
|-her2
...
Weight & Biases can be used to track experiments. It is turned off by default, but can be turned on by changing option in the config file in ./config/sabdab.yaml
@article {Ucar2024.10.07.617023,
author = {Ucar, Talip and Malherbe, Cedric and Gonzalez Hernandez, Ferran},
title = {Exploring Log-Likelihood Scores for Ranking Antibody Sequence Designs},
elocation-id = {2024.10.07.617023},
year = {2024},
doi = {10.1101/2024.10.07.617023},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/10/24/2024.10.07.617023},
eprint = {https://www.biorxiv.org/content/early/2024/10/24/2024.10.07.617023.full.pdf},
journal = {bioRxiv}
}
If you use DiffAbXL in your own studies, and work, please cite it by using the following:
@Misc{talip_ucar_2024_DiffAbXL,
author = {Talip Ucar},
title = {Exploring Log-Likelihood Scores for Ranking Antibody Sequence Designs},
URL = {https://github.com/AstraZeneca/DiffAbXL},
month = {October},
year = {since 2024}
}