med seg diff pytorch
0.3.3
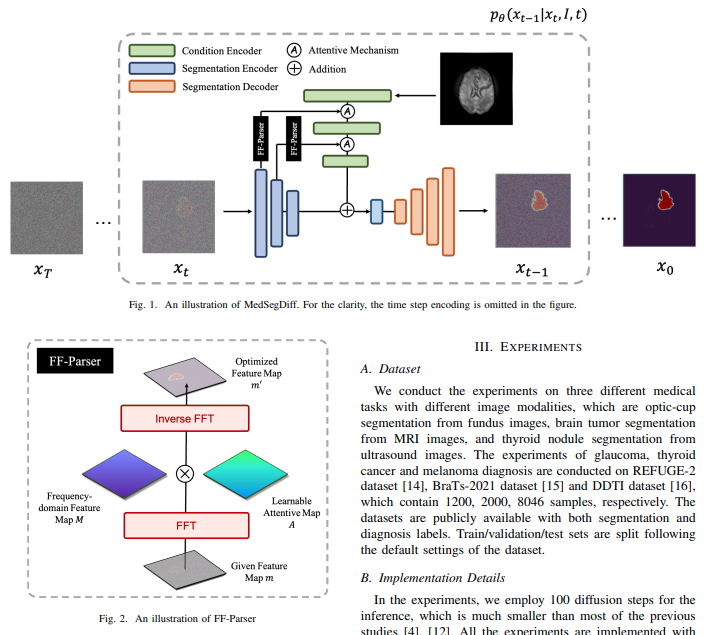
Implementation of MedSegDiff in Pytorch - SOTA medical segmentation out of Baidu using DDPM and enhanced conditioning on the feature level, with filtering of features in fourier space.
StabilityAI for the generous sponsorship, as well as my other sponsors out there
Isamu and Daniel for adding a training script for a skin lesion dataset!
$ pip install med-seg-diff-pytorchimport torch
from med_seg_diff_pytorch import Unet, MedSegDiff
model = Unet(
dim = 64,
image_size = 128,
mask_channels = 1, # segmentation has 1 channel
input_img_channels = 3, # input images have 3 channels
dim_mults = (1, 2, 4, 8)
)
diffusion = MedSegDiff(
model,
timesteps = 1000
).cuda()
segmented_imgs = torch.rand(8, 1, 128, 128) # inputs are normalized from 0 to 1
input_imgs = torch.rand(8, 3, 128, 128)
loss = diffusion(segmented_imgs, input_imgs)
loss.backward()
# after a lot of training
pred = diffusion.sample(input_imgs) # pass in your unsegmented images
pred.shape # predicted segmented images - (8, 3, 128, 128)Command to run
accelerate launch driver.py --mask_channels=1 --input_img_channels=3 --image_size=64 --data_path='./data' --dim=64 --epochs=100 --batch_size=1 --scale_lr --gradient_accumulation_steps=4If you want to add in self condition where we condition with the mask we have so far, do --self_condition
@article{Wu2022MedSegDiffMI,
title = {MedSegDiff: Medical Image Segmentation with Diffusion Probabilistic Model},
author = {Junde Wu and Huihui Fang and Yu Zhang and Yehui Yang and Yanwu Xu},
journal = {ArXiv},
year = {2022},
volume = {abs/2211.00611}
}@inproceedings{Hoogeboom2023simpleDE,
title = {simple diffusion: End-to-end diffusion for high resolution images},
author = {Emiel Hoogeboom and Jonathan Heek and Tim Salimans},
year = {2023}
}