AlphaFold3 (AF3), newly developed by the DeepMind team, has made breakthrough progress in the field of protein structure prediction. It can not only predict the structure of a single protein, but also predict the structure of protein complexes, nucleic acids or small molecules. Downcodes editors will give you an in-depth understanding of the inner workings of AF3 and how it achieves this complex task through clever architecture. AF3 is like a skilled "baker" who accurately "bakes" the three-dimensional structure of the protein according to the provided "recipe" (protein sequence). The process is like drawing a fine painting, with layers superimposed and finally presented. out the complete structure.
AlphaFold3, referred to as AF3, is the latest masterpiece of the DeepMind team in the field of protein structure prediction. It is able to predict the structure not only of individual protein sequences but also of protein complexes, nucleic acids or small molecules. It's like if you give AF3 a "recipe" for a protein, it can "bake" the three-dimensional structure of the protein.
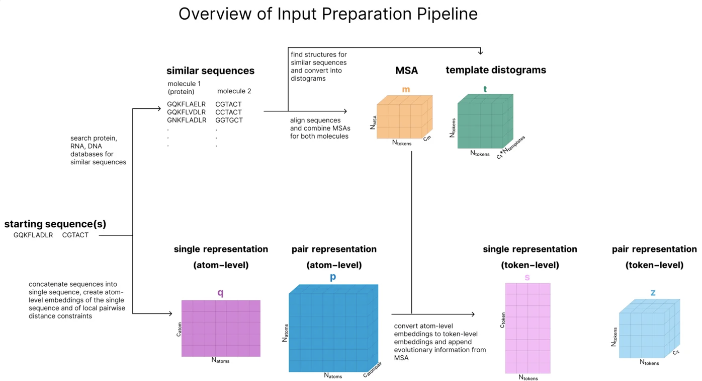
The architecture of AF3 is complex and subtle, but don’t be afraid, a picture can help you sort it out. The entire model can be divided into three parts:
Input preparation: Convert protein sequences into numerical tensors and retrieve molecules with similar structures.
Representation learning: Utilizing multiple attention mechanisms to update these representations.
Structure Prediction: Predict protein structures using conditional diffusion models.
Each step is like drawing a delicate painting. AF3 finally displays the three-dimensional structure of the protein through layer by layer.
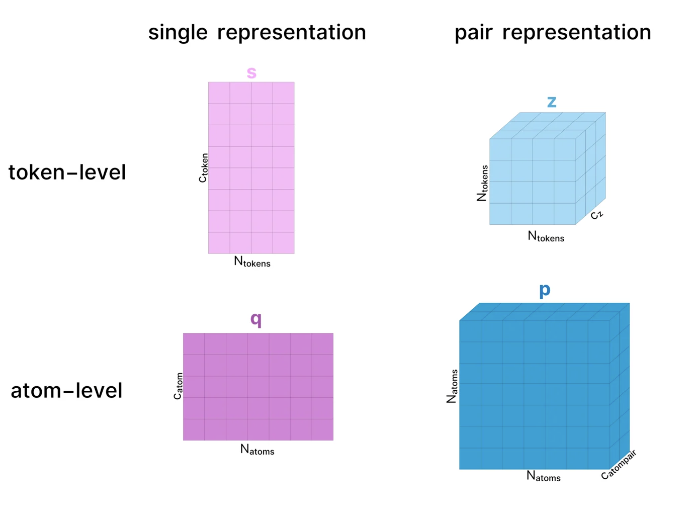
In the world of AF3, every molecule has its own "language". Whether it's proteins, DNA, RNA or small molecules, AF3 can convert them into a series of numerical tensors. It's like giving each molecule a unique "ID" that allows AF3 to recognize and process them.
The representation learning part of AF3 is like a carefully choreographed dance. Through the attention mechanism, AF3 can let the model's "eyesight" wander between different parts of the molecule and capture the relationship between them. This includes not only interactions within molecules, but also interactions between molecules.
In the structure prediction part of AF3, the conditional diffusion model plays a key role. It starts with a series of random noise and gradually "denoises" it, eventually restoring the true structure of the protein. This process is like gradually revealing the hidden truth from a fog.
AF3's training involves a variety of loss functions and confidence heads, which work together to allow AF3 to predict structures more accurately and evaluate how reliable its predictions are. It's like putting a mirror on AF3, allowing it to reflect and improve itself.
Reference: https://elanapearl.github.io/blog/2024/the-illustrated-alphafold/
All in all, AlphaFold3 has brought revolutionary changes to the field of protein structure prediction with its exquisite architecture and powerful learning capabilities. It has broad application prospects and is expected to play a huge role in biomedicine, materials science and other fields. I hope the explanation by the editor of Downcodes can help you better understand this amazing technology.