Early detection of cancer is key to improving patient survival, but existing technologies are challenged by insufficient sensitivity and specificity. Many methods rely on invasive examinations or have high false positive rates, limiting their widespread application. In recent years, liquid biopsy technology has attracted much attention due to its non-invasiveness, but its limitations in deep targeted sequencing have also restricted its development.
Early detection is always a tricky issue in the cancer treatment journey. In recent years, liquid biopsy technology has received widespread attention due to its non-invasiveness and high sensitivity. However, existing detection technologies mostly rely on deep targeted sequencing and are difficult to integrate multiple data types, thus affecting sensitivity and specificity.
In response to this technical shortcoming, a research team from the University of Oxford developed a new multi-modal circulating tumor DNA (ctDNA) detection method based on genome-wide TET-assisted pyridine borane sequencing (TAPS). The biggest highlight of this method is that it can analyze genome and methylation data simultaneously, making the sensitivity of cancer diagnosis reach 94.9% and the specificity reach 88.8%. This breakthrough technology provides new possibilities for early cancer screening and patient stratification.
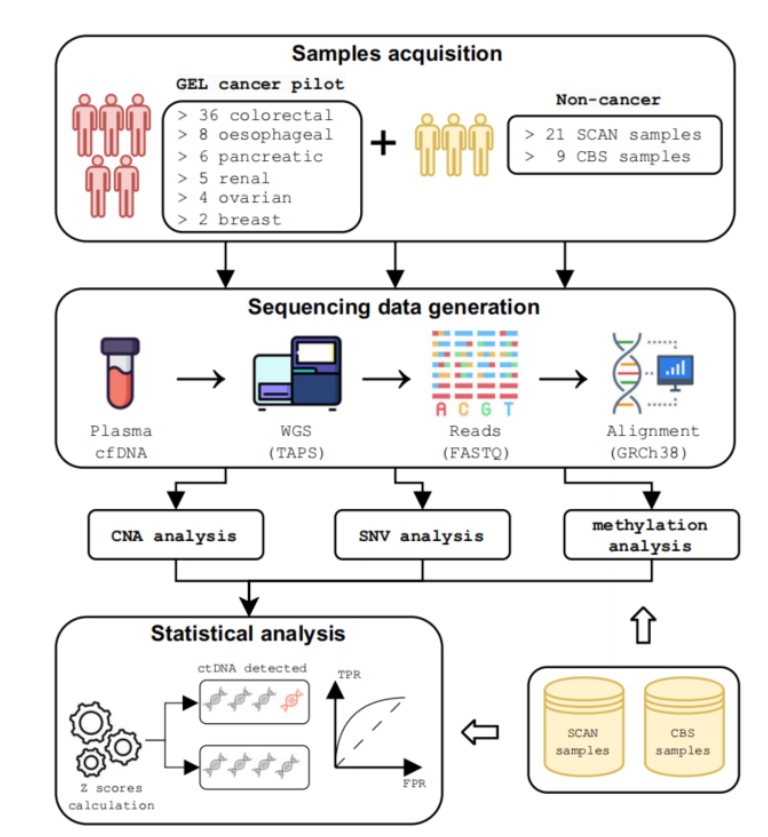
The study was titled "Multimodal cell-free DNA whole-genome TAPS is sensitive and reveals specific cancer signals" and was published in the journal "Nature Communications" on January 8, 2025. Research background shows that although early detection of cancer is crucial to improving patient prognosis, current screening methods can only cover less than 30% of cancer types, and many methods require invasive examinations and have low acceptance. Although early detection technology for multiple cancers can achieve non-invasive detection, the false positive rate is often high in asymptomatic people, which limits its application.
The Oxford team's TAPS technology uses a non-destructive method to maintain high sensitivity at low ctDNA content. The researchers verified the accuracy of this method in multiple cancer types by deep sequencing samples from 61 cancer patients and 30 non-cancer controls.
The team also developed a multimodal data analysis pipeline that integrates copy number variations, somatic mutations, and methylation signals to improve the sensitivity of ctDNA detection. The results showed that in clinical samples, the detection sensitivity of this method reached 85.2%, which was much higher than the results of a single data modality.
Although this method has shown significant advantages in early detection of cancer and postoperative monitoring, it still faces challenges in practical application, such as high sequencing costs and resource-limited clinical settings. Future studies can further optimize the sequencing technology to expand its applicability to more cancer types.
This research by the Oxford University team brings new hope for early detection of cancer, and its high sensitivity and specificity provide a solid foundation for more precise cancer diagnosis and treatment in the future. Although challenges remain, advances in this technology will undoubtedly advance the field of cancer treatment and bring benefits to more patients.