ANCOM Code Archive
Third release of ANCOM
请注意,此存储库仅用于存档目的。此存储库中的独立 ANCOM 功能不再维护。强烈建议用户使用我们的 ANCOMBC R 包中的ancom或ancombc函数。有关错误报告,请访问 ANCOMBC 存储库。
当前代码在横截面和纵向数据集中实现 ANCOM,同时允许使用协变量。需要包含以下库才能运行 R 代码:
library( nlme )
library( tidyverse )
library( compositions )
source( " programs/ancom.R " )我们采用ANCOM-II的方法作为预处理步骤,在进行差异丰度分析之前处理不同类型的零。
feature_table_pre_process(feature_table, meta_data, sample_var, group_var = NULL, out_cut = 0.05, zero_cut = 0.90, lib_cut, neg_lb)feature_table :表示观察到的 OTU/SV 表的数据框或矩阵,其中分类单元在行 ( rownames ) 中,样本在列 ( colnames ) 中。请注意,这是绝对丰度表,请勿将其转换为相对丰度表(其中列总计等于 1)。meta_data :所有感兴趣的变量和协变量的数据框或矩阵。sample_var :字符。存储样本 ID 的列的名称。group_var :字符。组指示器的名称。 group_var是检测结构零点和异常值所必需的。不同零点(结构零点、离群点零点、抽样零点)的定义请参考ANCOM-II。out_cut 0 到 1 之间的数值分数。对于每个分类单元,混合分布比例小于out_cut的观测值将被检测为异常值零;而混合分布比例大于1 - out_cut的观测值将被检测为异常值。zero_cut :0 和 1 之间的数值分数。零比例大于zero_cut的分类群不包括在分析中。lib_cut :数字。文库大小小于lib_cut的样本不包含在分析中。neg_lb :逻辑。 TRUE 表示分类单元将使用其渐近下界被分类为相应实验组中的结构零。更具体地说, neg_lb = TRUE表示您正在使用 ANCOM-II 第 3.2 节中所述的两个标准来检测结构零点;否则, neg_lb = FALSE将仅使用 ANCOM-II 第 3.2 节中的方程 1 来声明结构零。 feature_table :预处理OTU表的数据帧。meta_data :预处理元数据的数据框。structure_zeros :由 0 和 1 组成的矩阵,其中 1 表示该分类单元被识别为相应组中的结构零。ANCOM(feature_table, meta_data, struc_zero, main_var, p_adj_method, alpha, adj_formula, rand_formula, lme_control)feature_table :表示 OTU/SV 表的数据框,行中包含分类单元 ( rownames ),列中包含样本 ( colnames )。它可以是feature_table_pre_process的输出值。请注意,这是绝对丰度表,请勿将其转换为相对丰度表(其中列总计等于 1)。meta_data :变量的数据框。可以是feature_table_pre_process的输出值。struc_zero :由 0 和 1 组成的矩阵,其中 1 表示该分类单元被识别为相应组中的结构零。可以是feature_table_pre_process的输出值。main_var :字符。主要感兴趣变量的名称。 ANCOM v2.1 目前支持分类main_var 。p_adjust_method :字符。指定调整多重比较 p 值的方法。默认值为“BH”(Benjamini-Hochberg 过程)。alpha :显着性级别。默认值为 0.05。adj_formula :表示调整公式的字符串(参见示例)。rand_formula :表示lme中随机效应公式的字符串。有关详细信息,请参阅?lme 。lme_control :指定 lme 拟合控制值的列表。有关详细信息,请参阅?lmeControl 。 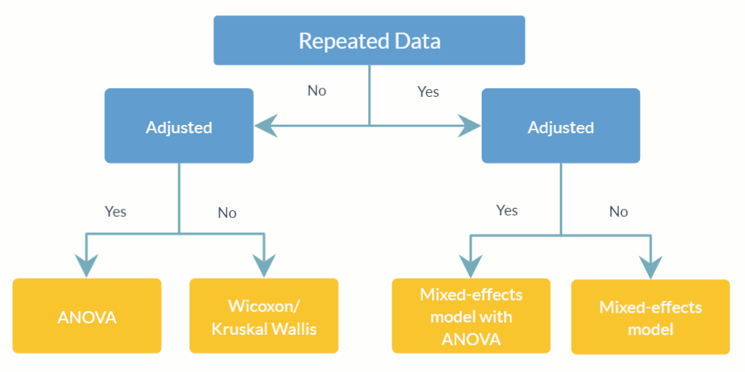
out :包含每个分类群和后续列的W统计量的数据框,这些列是 OTU 或分类群在一系列截止值(0.9、0.8、0.7 和 0.6)下是否具有差异丰度的逻辑指标。常用的是detected_0.7 。但是,如果您想对结果更加保守(较小的 FDR),则可以选择detected_0.8或detected_0.9 ,或者如果您想探索更多发现(较大的功效),则可以使用detected_0.6
fig :火山图的ggplot对象。
library( readr )
library( tidyverse )
otu_data = read_tsv( " data/moving-pics-table.tsv " , skip = 1 )
otu_id = otu_data $ `feature-id`
otu_data = data.frame ( otu_data [, - 1 ], check.names = FALSE )
rownames( otu_data ) = otu_id
meta_data = read_tsv( " data/moving-pics-sample-metadata.tsv " )[ - 1 , ]
meta_data = meta_data % > %
rename( Sample.ID = SampleID )
source( " programs/ancom.R " )
# Step 1: Data preprocessing
feature_table = otu_data ; sample_var = " Sample.ID " ; group_var = NULL
out_cut = 0.05 ; zero_cut = 0.90 ; lib_cut = 1000 ; neg_lb = FALSE
prepro = feature_table_pre_process( feature_table , meta_data , sample_var , group_var ,
out_cut , zero_cut , lib_cut , neg_lb )
feature_table = prepro $ feature_table # Preprocessed feature table
meta_data = prepro $ meta_data # Preprocessed metadata
struc_zero = prepro $ structure_zeros # Structural zero info
# Step 2: ANCOM
main_var = " Subject " ; p_adj_method = " BH " ; alpha = 0.05
adj_formula = NULL ; rand_formula = NULL ; lme_control = NULL
res = ANCOM( feature_table , meta_data , struc_zero , main_var , p_adj_method ,
alpha , adj_formula , rand_formula , lme_control )
write_csv( res $ out , " outputs/res_moving_pics.csv " )
# Step 3: Volcano Plot
# Number of taxa except structural zeros
n_taxa = ifelse(is.null( struc_zero ), nrow( feature_table ), sum(apply( struc_zero , 1 , sum ) == 0 ))
# Cutoff values for declaring differentially abundant taxa
cut_off = c( 0.9 * ( n_taxa - 1 ), 0.8 * ( n_taxa - 1 ), 0.7 * ( n_taxa - 1 ), 0.6 * ( n_taxa - 1 ))
names( cut_off ) = c( " detected_0.9 " , " detected_0.8 " , " detected_0.7 " , " detected_0.6 " )
# Annotation data
dat_ann = data.frame ( x = min( res $ fig $ data $ x ), y = cut_off [ " detected_0.7 " ], label = " W[0.7] " )
fig = res $ fig +
geom_hline( yintercept = cut_off [ " detected_0.7 " ], linetype = " dashed " ) +
geom_text( data = dat_ann , aes( x = x , y = y , label = label ),
size = 4 , vjust = - 0.5 , hjust = 0 , color = " orange " , parse = TRUE )
fig library( readr )
library( tidyverse )
otu_data = read_tsv( " data/moving-pics-table.tsv " , skip = 1 )
otu_id = otu_data $ `feature-id`
otu_data = data.frame ( otu_data [, - 1 ], check.names = FALSE )
rownames( otu_data ) = otu_id
meta_data = read_tsv( " data/moving-pics-sample-metadata.tsv " )[ - 1 , ]
meta_data = meta_data % > %
rename( Sample.ID = SampleID )
source( " programs/ancom.R " )
# Step 1: Data preprocessing
feature_table = otu_data ; sample_var = " Sample.ID " ; group_var = NULL
out_cut = 0.05 ; zero_cut = 0.90 ; lib_cut = 1000 ; neg_lb = FALSE
prepro = feature_table_pre_process( feature_table , meta_data , sample_var , group_var ,
out_cut , zero_cut , lib_cut , neg_lb )
feature_table = prepro $ feature_table # Preprocessed feature table
meta_data = prepro $ meta_data # Preprocessed metadata
struc_zero = prepro $ structure_zeros # Structural zero info
# Step 2: ANCOM
main_var = " Subject " ; p_adj_method = " BH " ; alpha = 0.05
adj_formula = " ReportedAntibioticUsage " ; rand_formula = NULL ; lme_control = NULL
res = ANCOM( feature_table , meta_data , struc_zero , main_var , p_adj_method ,
alpha , adj_formula , rand_formula , lme_control )group_var来识别结构零。在这里我们想知道不同delivery水平之间是否存在一些结构性零点library( readr )
library( tidyverse )
otu_data = read_tsv( " data/ecam-table-taxa.tsv " , skip = 1 )
otu_id = otu_data $ `feature-id`
otu_data = data.frame ( otu_data [, - 1 ], check.names = FALSE )
rownames( otu_data ) = otu_id
meta_data = read_tsv( " data/ecam-sample-metadata.tsv " )[ - 1 , ]
meta_data = meta_data % > %
rename( Sample.ID = `#SampleID` ) % > %
mutate( month = as.numeric( month ))
source( " programs/ancom.R " )
# Step 1: Data preprocessing
feature_table = otu_data ; sample_var = " Sample.ID " ; group_var = " delivery "
out_cut = 0.05 ; zero_cut = 0.90 ; lib_cut = 0 ; neg_lb = TRUE
prepro = feature_table_pre_process( feature_table , meta_data , sample_var , group_var ,
out_cut , zero_cut , lib_cut , neg_lb )
feature_table = prepro $ feature_table # Preprocessed feature table
meta_data = prepro $ meta_data # Preprocessed metadata
struc_zero = prepro $ structure_zeros # Structural zero info
# Step 2: ANCOM
main_var = " delivery " ; p_adj_method = " BH " ; alpha = 0.05
adj_formula = NULL ; rand_formula = " ~ 1 | studyid "
lme_control = list ( maxIter = 100 , msMaxIter = 100 , opt = " optim " )
res = ANCOM( feature_table , meta_data , struc_zero , main_var , p_adj_method ,
alpha , adj_formula , rand_formula , lme_control )
write_csv( res $ out , " outputs/res_ecam.csv " )
# Step 3: Volcano Plot
# Number of taxa except structural zeros
n_taxa = ifelse(is.null( struc_zero ), nrow( feature_table ), sum(apply( struc_zero , 1 , sum ) == 0 ))
# Cutoff values for declaring differentially abundant taxa
cut_off = c( 0.9 * ( n_taxa - 1 ), 0.8 * ( n_taxa - 1 ), 0.7 * ( n_taxa - 1 ), 0.6 * ( n_taxa - 1 ))
names( cut_off ) = c( " detected_0.9 " , " detected_0.8 " , " detected_0.7 " , " detected_0.6 " )
# Annotation data
dat_ann = data.frame ( x = min( res $ fig $ data $ x ), y = cut_off [ " detected_0.7 " ], label = " W[0.7] " )
fig = res $ fig +
geom_hline( yintercept = cut_off [ " detected_0.7 " ], linetype = " dashed " ) +
geom_text( data = dat_ann , aes( x = x , y = y , label = label ),
size = 4 , vjust = - 0.5 , hjust = 0 , color = " orange " , parse = TRUE )
fig group_var来识别结构零。在这里我们想知道不同delivery水平之间是否存在一些结构性零点library( readr )
library( tidyverse )
otu_data = read_tsv( " data/ecam-table-taxa.tsv " , skip = 1 )
otu_id = otu_data $ `feature-id`
otu_data = data.frame ( otu_data [, - 1 ], check.names = FALSE )
rownames( otu_data ) = otu_id
meta_data = read_tsv( " data/ecam-sample-metadata.tsv " )[ - 1 , ]
meta_data = meta_data % > %
rename( Sample.ID = `#SampleID` ) % > %
mutate( month = as.numeric( month ))
source( " programs/ancom.R " )
# Step 1: Data preprocessing
feature_table = otu_data ; sample_var = " Sample.ID " ; group_var = " delivery "
out_cut = 0.05 ; zero_cut = 0.90 ; lib_cut = 0 ; neg_lb = TRUE
prepro = feature_table_pre_process( feature_table , meta_data , sample_var , group_var ,
out_cut , zero_cut , lib_cut , neg_lb )
feature_table = prepro $ feature_table # Preprocessed feature table
meta_data = prepro $ meta_data # Preprocessed metadata
struc_zero = prepro $ structure_zeros # Structural zero info
# Step 2: ANCOM
main_var = " delivery " ; p_adj_method = " BH " ; alpha = 0.05
adj_formula = " month " ; rand_formula = " ~ 1 | studyid "
lme_control = list ( maxIter = 100 , msMaxIter = 100 , opt = " optim " )
res = ANCOM( feature_table , meta_data , struc_zero , main_var , p_adj_method ,
alpha , adj_formula , rand_formula , lme_control )group_var来识别结构零。在这里我们想知道不同级别的delivery之间是否存在一些结构性零点library( readr )
library( tidyverse )
otu_data = read_tsv( " data/ecam-table-taxa.tsv " , skip = 1 )
otu_id = otu_data $ `feature-id`
otu_data = data.frame ( otu_data [, - 1 ], check.names = FALSE )
rownames( otu_data ) = otu_id
meta_data = read_tsv( " data/ecam-sample-metadata.tsv " )[ - 1 , ]
meta_data = meta_data % > %
rename( Sample.ID = `#SampleID` ) % > %
mutate( month = as.numeric( month ))
source( " programs/ancom.R " )
# Step 1: Data preprocessing
feature_table = otu_data ; sample_var = " Sample.ID " ; group_var = " delivery "
out_cut = 0.05 ; zero_cut = 0.90 ; lib_cut = 0 ; neg_lb = TRUE
prepro = feature_table_pre_process( feature_table , meta_data , sample_var , group_var ,
out_cut , zero_cut , lib_cut , neg_lb )
feature_table = prepro $ feature_table # Preprocessed feature table
meta_data = prepro $ meta_data # Preprocessed metadata
struc_zero = prepro $ structure_zeros # Structural zero info
# Step 2: ANCOM
main_var = " delivery " ; p_adj_method = " BH " ; alpha = 0.05
adj_formula = " month " ; rand_formula = " ~ month | studyid "
lme_control = list ( maxIter = 100 , msMaxIter = 100 , opt = " optim " )
res = ANCOM( feature_table , meta_data , struc_zero , main_var , p_adj_method ,
alpha , adj_formula , rand_formula , lme_control )