BigWig 文件的快速批量数据加载,包含由 GPU 支持的表观轨迹数据和相应序列,用于深度学习应用。
Bigwig-loader主要依赖rapidsai kvikio库和cupy,这两个库最好使用conda/mamba安装。 Bigwig-loader 现在也可以使用 conda/mamba 安装。要创建安装了 bigwig-loader 的新环境:
mamba create -n my-env -c rapidsai -c conda-forge -c bioconda -c dataloading bigwig-loader或者将其添加到您的environment.yml 文件中:
name : my-env
channels :
- rapidsai
- conda-forge
- bioconda
- dataloading
dependencies :
- bigwig-loader并更新:
mamba env update -f environment.ymlBigwig-loader 也可以在已经安装了rapidsai kvikio 库和cupy 的环境中使用pip 安装:
pip install bigwig-loader我们将 BigWigDataset 包装在 PyTorch 可迭代数据集中,您可以直接使用:
# examples/pytorch_example.py
import pandas as pd
import torch
from torch . utils . data import DataLoader
from bigwig_loader import config
from bigwig_loader . pytorch import PytorchBigWigDataset
from bigwig_loader . download_example_data import download_example_data
# Download example data to play with
download_example_data ()
example_bigwigs_directory = config . bigwig_dir
reference_genome_file = config . reference_genome
train_regions = pd . DataFrame ({ "chrom" : [ "chr1" , "chr2" ], "start" : [ 0 , 0 ], "end" : [ 1000000 , 1000000 ]})
dataset = PytorchBigWigDataset (
regions_of_interest = train_regions ,
collection = example_bigwigs_directory ,
reference_genome_path = reference_genome_file ,
sequence_length = 1000 ,
center_bin_to_predict = 500 ,
window_size = 1 ,
batch_size = 32 ,
super_batch_size = 1024 ,
batches_per_epoch = 20 ,
maximum_unknown_bases_fraction = 0.1 ,
sequence_encoder = "onehot" ,
n_threads = 4 ,
return_batch_objects = True ,
)
# Don't use num_workers > 0 in DataLoader. The heavy
# lifting/parallelism is done on cuda streams on the GPU.
dataloader = DataLoader ( dataset , num_workers = 0 , batch_size = None )
class MyTerribleModel ( torch . nn . Module ):
def __init__ ( self ):
super (). __init__ ()
self . linear = torch . nn . Linear ( 4 , 2 )
def forward ( self , batch ):
return self . linear ( batch ). transpose ( 1 , 2 )
model = MyTerribleModel ()
optimizer = torch . optim . SGD ( model . parameters (), lr = 0.01 )
def poisson_loss ( pred , target ):
return ( pred - target * torch . log ( pred . clamp ( min = 1e-8 ))). mean ()
for batch in dataloader :
# batch.sequences.shape = n_batch (32), sequence_length (1000), onehot encoding (4)
pred = model ( batch . sequences )
# batch.values.shape = n_batch (32), n_tracks (2) center_bin_to_predict (500)
loss = poisson_loss ( pred [:, :, 250 : 750 ], batch . values )
print ( loss )
optimizer . zero_grad ()
loss . backward ()
optimizer . step ()可以从bigwig_loader.dataset导入与框架无关的 Dataset 对象。该数据集对象返回 cupy 张量。 Cupy 张量遵循 cuda 数组接口,可以零复制转换为 JAX 或张量流张量。
from bigwig_loader . dataset import BigWigDataset
dataset = BigWigDataset (
regions_of_interest = train_regions ,
collection = example_bigwigs_directory ,
reference_genome_path = reference_genome_file ,
sequence_length = 1000 ,
center_bin_to_predict = 500 ,
window_size = 1 ,
batch_size = 32 ,
super_batch_size = 1024 ,
batches_per_epoch = 20 ,
maximum_unknown_bases_fraction = 0.1 ,
sequence_encoder = "onehot" ,
)有关更多示例,请参阅示例目录。
该库用于加载具有相同维度的批量数据,这允许一些可以加快加载过程的假设。从下图可以看出,当加载少量数据时,pyBigWig 速度非常快,但没有利用数据加载的批量特性进行机器学习。
在下面的基准测试中,我们还使用 pyBigWig 创建了 PyTorch 数据加载器(使用 set_start_method('spawn')),以与每个 GPU 使用多个 CPU 的实际场景进行比较。我们看到 CPU 数据加载器的吞吐量并不随 CPU 数量线性增加,因此很难获得所需的吞吐量来保持 GPU 训练神经网络在学习步骤期间饱和。
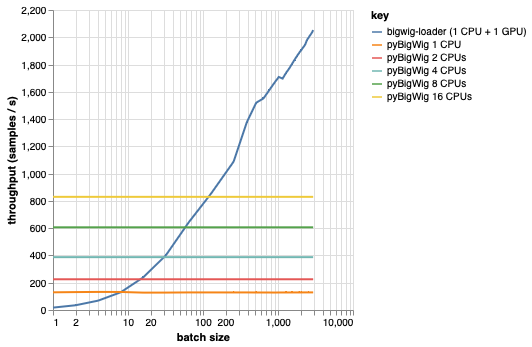
这就是bigwig-loader 解决的问题。这是如何使用 bigwig-loader 的示例:
git clone [email protected]:pfizer-opensource/bigwig-loadercd bigwig-loaderconda env create -f environment.yml在此环境中,您应该能够运行pytest -v并看到测试成功。注意:您需要 GPU 才能使用 bigwig-loader!
本部分将指导您完成添加新功能所需的步骤。如果有任何不清楚的地方,请提出问题。
git clone [email protected]:pfizer-opensource/bigwig-loadercd bigwig-loaderconda env create -f environment.ymlpip install -e '.[dev]'pre-commit install以安装预提交挂钩测试位于测试目录中。最重要的测试之一是 test_against_pybigwig,它确保如果 pyBigWIg 中存在错误,那么 bigwig-loader 中也会出现错误。
pytest -vv .当带有 GPU 的 github 运行程序可用时,我们还希望在 CI 中运行这些测试。但目前,您可以在本地运行它们。
如果您使用此库,请考虑引用:
雷特尔、乔伦·塞巴斯蒂安、安德烈亚斯·珀尔曼、乔什·邱、安德烈亚斯·斯特芬和德约克·阿内·克莱维特。 “用于 BigWig 文件表观遗传轨迹的快速机器学习数据加载器。”生物信息学 40,没有。 1(2024 年 1 月 1 日):btad767。 https://doi.org/10.1093/bioinformatics/btad767。
@article {
retel_fast_2024,
title = { A fast machine learning dataloader for epigenetic tracks from {BigWig} files } ,
volume = { 40 } ,
issn = { 1367-4811 } ,
url = { https://doi.org/10.1093/bioinformatics/btad767 } ,
doi = { 10.1093/bioinformatics/btad767 } ,
abstract = { We created bigwig-loader, a data-loader for epigenetic profiles from BigWig files that decompresses and processes information for multiple intervals from multiple BigWig files in parallel. This is an access pattern needed to create training batches for typical machine learning models on epigenetics data. Using a new codec, the decompression can be done on a graphical processing unit (GPU) making it fast enough to create the training batches during training, mitigating the need for saving preprocessed training examples to disk.The bigwig-loader installation instructions and source code can be accessed at https://github.com/pfizer-opensource/bigwig-loader } ,
number = { 1 } ,
urldate = { 2024-02-02 } ,
journal = { Bioinformatics } ,
author = { Retel, Joren Sebastian and Poehlmann, Andreas and Chiou, Josh and Steffen, Andreas and Clevert, Djork-Arné } ,
month = jan,
year = { 2024 } ,
pages = { btad767 } ,
}