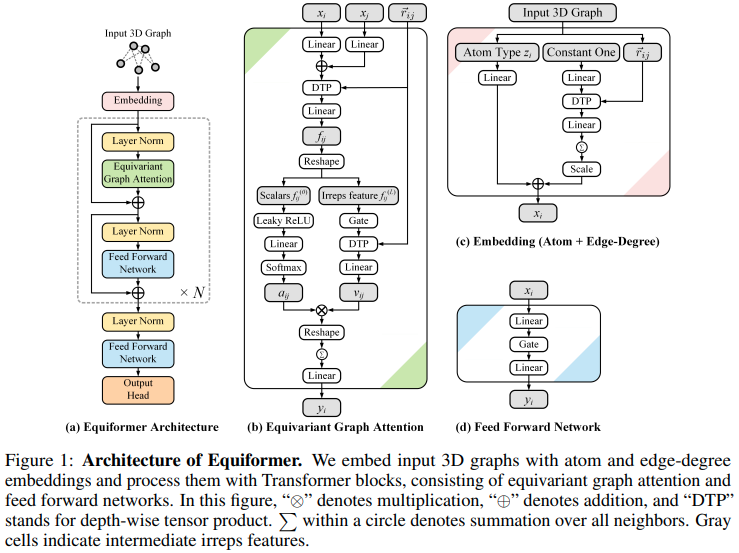
实现 Equiformer、SE3/E3 等变注意力网络,达到新的 SOTA,并被 EquiFold(Prescient Design)采用用于蛋白质折叠
这个设计似乎是建立在 SE3 Transformers 的基础上的,用 MLP Attention 和 GATv2 的非线性消息传递取代了点积注意力。它还进行深度张量乘积以提高效率。如果您认为我错了,请随时给我发电子邮件。
更新:有一项新的进展,使 SE3 等变网络的度数扩展变得更好!本文首先指出,通过沿 z 轴(或按其他惯例为 y 轴)对齐表示,球谐函数会变得稀疏。这从方程中删除了 m f维度。 Passaro 等人的后续论文。注意到 Clebsch Gordan 矩阵也变得稀疏,导致mi和 l f被删除。他们还认为,在将重复次数与一个轴对齐后,问题已从 SO(3) 减少到 SO(2)。 Equiformer v2(官方存储库)在类似 Transformer 的框架中利用这一点来达到新的 SOTA。
肯定会投入更多的工作/探索。现在,我已经将前两篇论文中的技巧融入到 Equiformer v1 中,除了完全转换为 SO(2) 之外。
更新 2:似乎有一个新的 SOTA,在更高程度的代表之间没有任何交互(换句话说,所有张量积/clebsch gordan 数学都消失了)。 GotenNet,似乎是 HEGNN 的变形版
$ pip install equiformer-pytorch import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 24 ,
dim = ( 4 , 4 , 2 ), # dimensions per type, ascending, length must match number of degrees (num_degrees)
dim_head = ( 4 , 4 , 4 ), # dimension per attention head
heads = ( 2 , 2 , 2 ), # number of attention heads
num_linear_attn_heads = 0 , # number of global linear attention heads, can see all the neighbors
num_degrees = 3 , # number of degrees
depth = 4 , # depth of equivariant transformer
attend_self = True , # attending to self or not
reduce_dim_out = True , # whether to reduce out to dimension of 1, say for predicting new coordinates for type 1 features
l2_dist_attention = False # set to False to try out MLP attention
). cuda ()
feats = torch . randint ( 0 , 24 , ( 1 , 128 )). cuda ()
coors = torch . randn ( 1 , 128 , 3 ). cuda ()
mask = torch . ones ( 1 , 128 ). bool (). cuda ()
out = model ( feats , coors , mask ) # (1, 128)
out . type0 # invariant type 0 - (1, 128)
out . type1 # equivariant type 1 - (1, 128, 3)该存储库还包括一种使用可逆网络将内存使用与深度解耦的方法。换句话说,如果增加深度,则使用一个等量变换器块(注意力和前馈)时,内存成本将保持不变。
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 24 ,
dim = ( 4 , 4 , 2 ),
dim_head = ( 4 , 4 , 4 ),
heads = ( 2 , 2 , 2 ),
num_degrees = 3 ,
depth = 48 , # depth of 48 - just to show that it runs - in reality, seems to be quite unstable at higher depths, so architecture stil needs more work
reversible = True , # just set this to True to use https://arxiv.org/abs/1707.04585
). cuda ()
feats = torch . randint ( 0 , 24 , ( 1 , 128 )). cuda ()
coors = torch . randn ( 1 , 128 , 3 ). cuda ()
mask = torch . ones ( 1 , 128 ). bool (). cuda ()
out = model ( feats , coors , mask )
out . type0 . sum (). backward ()有边缘,例如。原子键
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 28 ,
dim = 64 ,
num_edge_tokens = 4 , # number of edge type, say 4 bond types
edge_dim = 16 , # dimension of edge embedding
depth = 2 ,
input_degrees = 1 ,
num_degrees = 3 ,
reduce_dim_out = True
)
atoms = torch . randint ( 0 , 28 , ( 2 , 32 ))
bonds = torch . randint ( 0 , 4 , ( 2 , 32 , 32 ))
coors = torch . randn ( 2 , 32 , 3 )
mask = torch . ones ( 2 , 32 ). bool ()
out = model ( atoms , coors , mask , edges = bonds )
out . type0 # (2, 32)
out . type1 # (2, 32, 3)与邻接矩阵
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
dim = 32 ,
heads = 8 ,
depth = 1 ,
dim_head = 64 ,
num_degrees = 2 ,
valid_radius = 10 ,
reduce_dim_out = True ,
attend_sparse_neighbors = True , # this must be set to true, in which case it will assert that you pass in the adjacency matrix
num_neighbors = 0 , # if you set this to 0, it will only consider the connected neighbors as defined by the adjacency matrix. but if you set a value greater than 0, it will continue to fetch the closest points up to this many, excluding the ones already specified by the adjacency matrix
num_adj_degrees_embed = 2 , # this will derive the second degree connections and embed it correctly
max_sparse_neighbors = 8 # you can cap the number of neighbors, sampled from within your sparse set of neighbors as defined by the adjacency matrix, if specified
)
feats = torch . randn ( 1 , 128 , 32 )
coors = torch . randn ( 1 , 128 , 3 )
mask = torch . ones ( 1 , 128 ). bool ()
# placeholder adjacency matrix
# naively assuming the sequence is one long chain (128, 128)
i = torch . arange ( 128 )
adj_mat = ( i [:, None ] <= ( i [ None , :] + 1 )) & ( i [:, None ] >= ( i [ None , :] - 1 ))
out = model ( feats , coors , mask , adj_mat = adj_mat )
out . type0 # (1, 128)
out . type1 # (1, 128, 3) 等方差测试等
$ python setup.py test 首先安装sidechainnet
$ pip install sidechainnet然后运行蛋白质主干去噪任务
$ python denoise.py将 xi 和 xj 单独的项目和求和逻辑移至 Conv 类中
将自交互键/值生成移至Conv,修复自交互中的无池化问题
采用一种简单的方法来分割 DTP 输入程度的贡献
对于更高类型的点积注意力,请尝试欧几里德距离
考虑仅针对 type0 的所有邻居注意力层,使用线性注意力
整合球形通道论文中的新发现,随后是 so(3) -> so(2) 论文,这减少了 O(L^6) -> O(L^3) 的计算!
@article { Liao2022EquiformerEG ,
title = { Equiformer: Equivariant Graph Attention Transformer for 3D Atomistic Graphs } ,
author = { Yi Liao and Tess E. Smidt } ,
journal = { ArXiv } ,
year = { 2022 } ,
volume = { abs/2206.11990 }
} @article { Lee2022.10.07.511322 ,
author = { Lee, Jae Hyeon and Yadollahpour, Payman and Watkins, Andrew and Frey, Nathan C. and Leaver-Fay, Andrew and Ra, Stephen and Cho, Kyunghyun and Gligorijevic, Vladimir and Regev, Aviv and Bonneau, Richard } ,
title = { EquiFold: Protein Structure Prediction with a Novel Coarse-Grained Structure Representation } ,
elocation-id = { 2022.10.07.511322 } ,
year = { 2022 } ,
doi = { 10.1101/2022.10.07.511322 } ,
publisher = { Cold Spring Harbor Laboratory } ,
URL = { https://www.biorxiv.org/content/early/2022/10/08/2022.10.07.511322 } ,
eprint = { https://www.biorxiv.org/content/early/2022/10/08/2022.10.07.511322.full.pdf } ,
journal = { bioRxiv }
} @article { Shazeer2019FastTD ,
title = { Fast Transformer Decoding: One Write-Head is All You Need } ,
author = { Noam M. Shazeer } ,
journal = { ArXiv } ,
year = { 2019 } ,
volume = { abs/1911.02150 }
} @misc { ding2021cogview ,
title = { CogView: Mastering Text-to-Image Generation via Transformers } ,
author = { Ming Ding and Zhuoyi Yang and Wenyi Hong and Wendi Zheng and Chang Zhou and Da Yin and Junyang Lin and Xu Zou and Zhou Shao and Hongxia Yang and Jie Tang } ,
year = { 2021 } ,
eprint = { 2105.13290 } ,
archivePrefix = { arXiv } ,
primaryClass = { cs.CV }
} @inproceedings { Kim2020TheLC ,
title = { The Lipschitz Constant of Self-Attention } ,
author = { Hyunjik Kim and George Papamakarios and Andriy Mnih } ,
booktitle = { International Conference on Machine Learning } ,
year = { 2020 }
} @article { Zitnick2022SphericalCF ,
title = { Spherical Channels for Modeling Atomic Interactions } ,
author = { C. Lawrence Zitnick and Abhishek Das and Adeesh Kolluru and Janice Lan and Muhammed Shuaibi and Anuroop Sriram and Zachary W. Ulissi and Brandon C. Wood } ,
journal = { ArXiv } ,
year = { 2022 } ,
volume = { abs/2206.14331 }
} @article { Passaro2023ReducingSC ,
title = { Reducing SO(3) Convolutions to SO(2) for Efficient Equivariant GNNs } ,
author = { Saro Passaro and C. Lawrence Zitnick } ,
journal = { ArXiv } ,
year = { 2023 } ,
volume = { abs/2302.03655 }
} @inproceedings { Gomez2017TheRR ,
title = { The Reversible Residual Network: Backpropagation Without Storing Activations } ,
author = { Aidan N. Gomez and Mengye Ren and Raquel Urtasun and Roger Baker Grosse } ,
booktitle = { NIPS } ,
year = { 2017 }
} @article { Bondarenko2023QuantizableTR ,
title = { Quantizable Transformers: Removing Outliers by Helping Attention Heads Do Nothing } ,
author = { Yelysei Bondarenko and Markus Nagel and Tijmen Blankevoort } ,
journal = { ArXiv } ,
year = { 2023 } ,
volume = { abs/2306.12929 } ,
url = { https://api.semanticscholar.org/CorpusID:259224568 }
} @inproceedings { Arora2023ZoologyMA ,
title = { Zoology: Measuring and Improving Recall in Efficient Language Models } ,
author = { Simran Arora and Sabri Eyuboglu and Aman Timalsina and Isys Johnson and Michael Poli and James Zou and Atri Rudra and Christopher R'e } ,
year = { 2023 } ,
url = { https://api.semanticscholar.org/CorpusID:266149332 }
}