这是Braker上首次BGA23研讨会会议的录音。如果通过观看视频来学习很容易,请考虑观看:https://www.youtube.com/watch?v=uxtkj4mukyg
Braker3现在在https://usegalaxy.eu/中
tsebra&braker3相关:
Braker&Augustus相关:
基因标记相关:
Mark Borodovsky,美国佐治亚理工学院,[email protected]
美国联合基因组学院Tomas Bruna,[email protected]
美国佐治亚理工学院的Alexandre Lomsazde,[email protected]
[A] Greifswald大学,数学与计算机科学研究所,Walther-Rathenau-STR。 47,17489德国格里夫斯瓦尔德
[B] Greifswald大学,Microbes功能基因组学中心,Felix-Hausdorff-STR。 8,17489德国格里夫斯瓦尔德
[C]佐治亚理工学院联合和埃默里大学Wallace H Coulter生物医学工程系,美国亚特兰大30332
[D]计算科学与工程学院,美国亚特兰大30332
[e]莫斯科物理技术研究所,莫斯科地区141701,俄罗斯多尔格普德尼
![Braker2-Team-2 [FIG10]](https://images.downcodes.com/uploads/20250214/img_67aee79a0cb7530.png)
![Braker2-Team-1 [FIG11]](https://images.downcodes.com/uploads/20250214/img_67aee79a0d1eb31.png)
![Braker2-Team-3 [FIG12]](https://images.downcodes.com/uploads/20250214/img_67aee79a0da9c32.png)
![Braker2-Team-4 [FIG13]](https://images.downcodes.com/uploads/20250214/img_67aee79a0e49933.png)
图1:当前的布拉克作者,从左到右:马里奥·斯坦克,亚历山大·洛姆德兹,凯塔琳娜·J·霍夫,托马斯·布鲁纳,拉尔斯·加布里埃尔和马克·博罗多夫斯基。我们承认,一个更大的科学家社区为布拉克守则做出了贡献(例如,通过拉动请求)。
BRAKER1,BRAKER2和BRAKER3的开发得到了美国国立卫生研究院(NIH)[GM128145 to MB和MS]的支持。 Braker3的开发部分由德国梅克伦堡 - Vorpommern政府授予KJH和MS的项目数据能力部分资助。
Braker(TSEBRA)的成绩单选择器可在https://github.com/gaius-augustus/tsebra上找到。
Genemark-Etp是Braker核心的基因发现者之一,可在https://github.com/gatech-genemark/genemark/genemark-etp上获得。
奥古斯都(Augustus)是布拉克(Braker)核心的第二个基因发现者,可在https://github.com/gaius-augustus/augustus上获得。
Galba是用于使用Miniprot或GenoMeThreader生成训练基因的Braker管道衍生产品,可在https://github.com/gaius-augustus/galba上获得。
快速增长的测序基因组需要完全自动化的方法才能准确基因结构注释。考虑到这个目标,我们开发了Braker1 R1 R0 ,这是Genemark-Et R2和Augustus R3 (R4)的组合,它使用基因组和RNA-SEQ数据自动在新型基因组中自动生成完整的基因结构注释。
但是,可用于注释新基因组的RNA-seq数据的质量是可变的,在某些情况下,RNA-Seq数据根本不可用。
Braker2是Braker1的扩展,它允许对基因预测工具进行完全自动化的培训, ET/EP/EP/EP/ETP R14, R15, R17, F1和Augustus,从RNA-Seq和/或蛋白质同源性信息中进行了整合,并整合了整合RNA-SEQ和蛋白质同源性信息的外在证据到预测。
与依赖蛋白质同源性信息的其他可用方法相反,即使没有非常紧密相关的物种的注释,也没有RNA-SEQ数据,Braker2即使在没有注释的情况下也达到了高基因预测准确性。
Braker3是Braker Suite的最新管道。它可以在完全自动化的管道中使用RNA-seq和蛋白质数据,以训练和预测具有Genemark-Etp和Augustus的高度可靠基因。管道的结果是两个基因预测工具的组合基因集,该基因仅包含外在证据的很高支持的基因。
在本用户指南中,我们将简单地参考Braker1,Braker2和Braker3作为Braker,因为它们由同一脚本执行( braker.pl )。
使用高质量的基因组组件。如果您的基因组组件中有大量短的脚手架,那么这些短脚手架可能会大大增加运行时,但不会提高预测准确性。
在基因组文件中使用简单的脚手架名称(例如>contig1比>contig1my custom species namesome putative function /more/information/ and lots of special characters %&!*(){} )。在运行任何对齐程序之前,请在所有Fasta文件中简单地制作脚手架名称。
为了准确预测新型基因组中的基因,应掩盖重复序列的基因组。这将避免重复和低复杂区域中假阳性基因结构的预测。重复掩蔽对于使用一些工具(其他RNA-seq映射器(例如HISAT2),忽略掩蔽信息)将RNA-Seq数据映射到基因组至关重要。对于Genemark-ES/ET/EP/ETP和Augustus的情况,软卸式(即将重复区域放入较低的案例字母和所有其他区域中的大写字母中)会导致比硬汉取得更好的结果(即通过字母在重复区域中替换重复区域的字母N用于未知核苷酸)。
许多基因组具有基因结构,这些基因结构将通过基因/ET/EP/ETP的标准参数进行准确预测,而布拉克内的奥古斯都将进行预测。但是,某些基因组具有特异性特异性特征,即真菌中的特殊分支点模型或非标准的剪接位点模式。请阅读选项部分[选项],以确定任何自定义选项是否可以提高目标物种基因组的基因预测准确性。
在进一步使用之前,请务必检查基因预测结果!您可以使用基因组浏览器在具有外在证据数据的上下文中目视检查基因模型。 Braker支持使用MakeHub为此目的为UCSC基因组浏览器的轨道数据集线器的生成。
Braker主要具有半无调的外部证据数据(RNA-SEQ和/或蛋白质剪接的对齐信息)支持基因公园/ET/EP/ETP [F1]的培训,以及随后对Augustus培训与最终的外部证据整合到最终的外部证据的培训基因预测步骤。但是,现在布拉克中还包括许多其他管道。在下文中,我们概述了可能的输入文件和管道:
![braker2-main-a [fig1]](https://images.downcodes.com/uploads/20250214/img_67aee79a0eaf534.png)
图2:Braker管道A:仅在基因组数据上训练基因标志;从头开始的基因预测
![Braker2-Main-B [FIG2]](https://images.downcodes.com/uploads/20250214/img_67aee79a0f13f35.png)
图3:Braker管道B:由RNA-seq剪接对准信息支持的训练Genemark-ET,与Augustus的预测以及相同的拼接比对信息进行了预测。
![Braker2-Main-C [FIF3]](https://images.downcodes.com/uploads/20250214/img_67aee79a0fa1036.png)
图4:Braker管道C:对蛋白质剪接比对,启动和停止信息的训练基因 - EP+与Augustus的预测相同的信息,此外,链接的CDSPART提示。这里使用的蛋白质可以是与目标生物的任何进化距离。
![Braker3-Main-A [FIF4]](https://images.downcodes.com/uploads/20250214/img_67aee79a1010b37.png)
图5:Braker管道D:如有必要,为目标物种下载和对齐RNA-Seq集。 RNA-seq比对支持和大蛋白数据库支持的基因标准-ETP(蛋白质可能是任何进化距离)。随后,奥古斯都使用相同的外部信息以及基因标准-ETP结果进行培训和预测。最终的预测是奥古斯都和基因标准结果的TSEBRA组合。
我们知道,Braker3及其所有依赖项的“手动”安装是乏味的,而且在没有根本权限的情况下确实具有挑战性。因此,我们提供了一个码头容器,该容器已开发为以奇异性运行。有关此容器的所有信息,请访问https://hub.docker.com/r/teambraker/braker3
简而言之,构建如下:
singularity build braker3.sif docker://teambraker/braker3:latest
执行:
singularity exec braker3.sif braker.pl
测试:
singularity exec -B $PWD:$PWD braker3.sif cp /opt/BRAKER/example/singularity-tests/test1.sh .
singularity exec -B $PWD:$PWD braker3.sif cp /opt/BRAKER/example/singularity-tests/test2.sh .
singularity exec -B $PWD:$PWD braker3.sif cp /opt/BRAKER/example/singularity-tests/test3.sh .
export BRAKER_SIF=/your/path/to/braker3.sif # may need to modify
bash test1.sh
bash test2.sh
bash test3.sh
很少有用户想在Docker内部进行分析(因为需要根本权限)。但是,如果这是您的目标,则可以运行并测试容器如下
sudo docker run --user 1000:100 --rm -it teambraker/braker3:latest bash
bash /opt/BRAKER/example/docker-tests/test1.sh # BRAKER1
bash /opt/BRAKER/example/docker-tests/test2.sh # BRAKER2
bash /opt/BRAKER/example/docker-tests/test3.sh # BRAKER3
祝你好运 ;-)
$PATH变量中徘徊的较旧的基因标记版可能会导致无法预料的干扰,从而导致程序失败。请将所有较旧的Genemark版本从您的$PATH中移出(例如,在ProtHint/dependencies中的基因)。
在发布时,该Braker版本进行了测试:
奥古斯都3.5.0 F2
Genemark-Etp(来源见Dockerfile)
BAMTools 2.5.1 R5
Samtools 1.7-4-G93586ED R6
Spaln 2.3.3d R8, R9, R10
NCBI BLAST+ 2.2.31+ R12, R13
钻石0.9.24
CDBFASTA 0.99
cdbyank 0.981
Gushr 1.0.0
SRA工具包3.00 R14
HISAT2 2.2.1 R15
BedTools 2.30 R16
StringTie2 2.2.1 R17
gffread 0.12.7 R18
ERPEASM 0.2.5 R27
运行Braker需要一个带有bash和Perl的Linux系统。此外,布拉克需要安装以下CPAN-Perl模块:
File::Spec::Functions
Hash::Merge
List::Util
MCE::Mutex
Module::Load::Conditional
Parallel::ForkManager
POSIX
Scalar::Util::Numeric
YAML
Math::Utils
File::HomeDir
对于当蛋白质和RNA-seq提供时使用的Genemark-ETP:
YAML::XSData::DumperThread::Queuethreads例如,在Ubuntu上,使用CPanminus F4安装模块: sudo cpanm Module::Name ,例如sudo cpanm Hash::Merge 。
Braker还使用CPAN上无法使用的Perl模块helpMod_braker.pm 。该模块是Braker版本的一部分,不需要单独的安装。
如果您在Linux计算机上没有根本权限,请尝试设置Anaconda (https://www.anaconda.com/distribution/)环境:
wget https://repo.anaconda.com/archive/Anaconda3-2018.12-Linux-x86_64.sh
bash bin/Anaconda3-2018.12-Linux-x86_64.sh # do not install VS (needs root privileges)
conda install -c anaconda perl
conda install -c anaconda biopython
conda install -c bioconda perl-app-cpanminus
conda install -c bioconda perl-file-spec
conda install -c bioconda perl-hash-merge
conda install -c bioconda perl-list-util
conda install -c bioconda perl-module-load-conditional
conda install -c bioconda perl-posix
conda install -c bioconda perl-file-homedir
conda install -c bioconda perl-parallel-forkmanager
conda install -c bioconda perl-scalar-util-numeric
conda install -c bioconda perl-yaml
conda install -c bioconda perl-class-data-inheritable
conda install -c bioconda perl-exception-class
conda install -c bioconda perl-test-pod
conda install -c bioconda perl-file-which # skip if you are not comparing to reference annotation
conda install -c bioconda perl-mce
conda install -c bioconda perl-threaded
conda install -c bioconda perl-list-util
conda install -c bioconda perl-math-utils
conda install -c bioconda cdbtools
conda install -c eumetsat perl-yaml-xs
conda install -c bioconda perl-data-dumper
随后在您的Conda环境中安装Braker和其他软件“照常”。注意:有一个Bioconda Braker套餐和一个Bioconda Augustus套餐。他们工作。但是它们通常落后于GitHub上这两个工具的开发代码。因此,我们建议您手动安装和最新来源的使用。
Braker是Perl和Python脚本和Perl模块的集合。为了运行Braker的主要脚本是braker.pl 。其他Perl和Python组件是:
align2hints.pl
filterGenemark.pl
filterIntronsFindStrand.pl
startAlign.pl
helpMod_braker.pm
findGenesInIntrons.pl
downsample_traingenes.pl
ensure_n_training_genes.py
get_gc_content.py
get_etp_hints.py
为了运行Braker,所有是Braker的一部分的脚本(以*.pl和*.py结尾的文件)必须可执行。如果您从Github下载Braker,则应该已经是这种情况。如果您在USB棒上转移Braker到另一台计算机,则可以覆盖可执行性。为了检查所需文件是否可执行,请在包含Braker Perl脚本的目录中运行以下命令:
ls -l *.pl *.py
输出应与此相似:
-rwxr-xr-x 1 katharina katharina 18191 Mai 7 10:25 align2hints.pl
-rwxr-xr-x 1 katharina katharina 6090 Feb 19 09:35 braker_cleanup.pl
-rwxr-xr-x 1 katharina katharina 408782 Aug 17 18:24 braker.pl
-rwxr-xr-x 1 katharina katharina 5024 Mai 7 10:25 downsample_traingenes.pl
-rwxr-xr-x 1 katharina katharina 5024 Mai 7 10:23 ensure_n_training_genes.py
-rwxr-xr-x 1 katharina katharina 4542 Apr 3 2019 filter_augustus_gff.pl
-rwxr-xr-x 1 katharina katharina 30453 Mai 7 10:25 filterGenemark.pl
-rwxr-xr-x 1 katharina katharina 5754 Mai 7 10:25 filterIntronsFindStrand.pl
-rwxr-xr-x 1 katharina katharina 7765 Mai 7 10:25 findGenesInIntrons.pl
-rwxr-xr-x 1 katharina katharina 1664 Feb 12 2019 gatech_pmp2hints.pl
-rwxr-xr-x 1 katharina katharina 2250 Jan 9 13:55 log_reg_prothints.pl
-rwxr-xr-x 1 katharina katharina 4679 Jan 9 13:55 merge_transcript_sets.pl
-rwxr-xr-x 1 katharina katharina 41674 Mai 7 10:25 startAlign.pl
重要的是,每个脚本都存在-rwxr-xr-x中的x 。如果不是这样,请运行
`chmod a+x *.pl *.py`
为了更改文件属性。
您可能会发现将Braker Perl脚本驻留到您的$PATH环境变量的目录很有帮助。对于一次bash会话,请输入:
PATH=/your_path_to_braker/:$PATH
export PATH
要使此$PATH修改可用于所有BASH会话,请在启动脚本(例如~/.bashrc )中添加上述行。
布雷克(Braker)呼吁不属于布拉克(Braker)的各种生物信息学软件工具。某些工具是必须的,即如果您的系统中不存在这些工具,则Braker根本不会运行。其他工具是可选的。请安装以您选择的方式运行Braker所需的所有工具。
从http://github.com/gatech-genemark/genemark-etp或https://topaz.gatech.gatech.edu/genemark/genemark/etp.for_braker.tar.gz下载genemark-etp f1 。按照Genemark-Etp的README文件中所述解开包装并安装Genemark-ETP。
如果已经包含在您的$PATH变量中,则Braker会自动猜测gmes_petap.pl或gmetp.pl的位置。否则,Braker可以通过在环境变量GENEMARK_PATH或使用命令行参数( --GENEMARK_PATH=/your_path_to_GeneMark_executables/ )中找到基因标记/EP/EP/ETP可执行文件。
为了为您的当前bash会话设置环境变量,请输入:
export GENEMARK_PATH=/your_path_to_GeneMark_executables/
将上面的行添加到启动脚本(例如~/.bashrc )中,以使其可用于所有bash会话。
Genemark-ES/ET/EP/ETP中的PERL脚本配置为/usr/bin/perl的默认Perl位置。
如果您在Anaconda环境中运行Genemark-es/et/EP/ETP(或出于任何其他原因,请使用$PATH变量中的Perl),请使用所有Genemark-es/et/ep/etp脚本的Shebang修改以下命令位于Genemark-ES/ET/EP/ETP文件夹中:
perl change_path_in_perl_scripts.pl "/usr/bin/env perl"
您可以通过运行check_install.bash和/或在GeneMark-E-tests目录中运行check_install.bash和/或执行示例来检查Genemark-ES/ET/EP是否正确安装。
Genemark-Etp是向下兼容的,即它也涵盖了Braker中Genemark-EP和Genemark-Et的功能。
从https://github.com/gaius-augustus/augustus上下载奥古斯都。根据Augustus README.TXT解开Augustus并安装Augustus。请勿使用其他来源的过时的奥古斯都版本,例如Debian软件包或Bioconda软件包!布雷克(Braker)高度依赖于最新的奥古斯都/脚本目录,其他来源通常落后。
您应该在自己的系统上编译奥古斯都,以避免奥古斯都使用的库版本问题。编译说明在Augustus README.TXT文件( Augustus/README.txt )中提供。
Augustus由augustus (基因预测工具)组成,位于Augustus/scripts中的Augustus/auxprogs和Perl脚本中的其他C ++工具。 Perl脚本必须是可执行的(请参阅Braker组件中的说明。
使用RNA-Seq运行时,C ++工具bam2hints是Braker的重要组成部分。来源位于Augustus/auxprogs/bam2hints中。确保您在系统上编译bam2hints (应该在编译Augustus时自动编译,但是如果bam2hints出现问题,请阅读Augustus/auxprogs/bam2hints/README中的故障排除说明)。
由于布雷克(Braker)是训练奥古斯都(Augustus)的管道,因此,IE写了特定的参数文件,因此布雷克(Braker)需要写入包含此类文件的奥古斯都(Augustus)配置目录( Augustus/config/ )。如果您在系统上全球安装Augustus,则配置config夹通常并非所有用户都写。将配置归于Augustus用户递归的config将目录制作,或将config/文件夹(递归)复制到用户具有写入许可的位置。
Augustus将通过查找环境变量$AUGUSTUS_CONFIG_PATH来找到config文件夹。如果未设置$AUGUSTUS_CONFIG_PATH环境变量,则Braker将在路径中查看../config相对于找到Augustus可执行文件的目录。另外,您可以将变量作为命令行参数提供给Braker( --AUGUSTUS_CONFIG_PATH=/your_path_to_AUGUSTUS/Augustus/config/ )。我们建议您在当前的bash会话中导出变量:
export AUGUSTUS_CONFIG_PATH=/your_path_to_AUGUSTUS/Augustus/config/
为了使变量可用于所有bash会话,请在启动脚本(例如~/.bashrc中添加上述行。
如果您想将Augustus作为Debian软件包,请查看Dockerfile。那么,需要修补许多脚本。
布拉克(Braker)期望奥古斯都(Augustus)的整个config目录$AUGUSTUS_CONFIG_PATH ,即具有其内容(至少generic )和extrinsic的子羊群species !以$AUGUSTUS_CONFIG_PATH提供可写的但空的文件夹将不适合Braker。如果您需要将Augustus二进制文件和$AUGUSTUS_CONFIG_PATH分开,我们建议您递归将不可4的配置内容复制到可写的位置。
如果您在/usr/bin/augustus上安装了Augustus的系统范围安装,则坐落在/usr/bin/augustus_config/的config副本。文件夹/home/yours/对您来说是可写的。使用以下命令复制(另外设置当时必需的变量):
cp -r /usr/bin/Augustus/config/ /home/yours/
export AUGUSTUS_CONFIG_PATH=/home/yours/augustus_config
export AUGUSTUS_BIN_PATH=/usr/bin
export AUGUSTUS_SCRIPTS_PATH=/usr/bin/augustus_scripts
将奥古斯都二进制文件和脚本的目录添加到您的$PATH变量中,使您的系统可以自动找到这些工具。这不是运行Braker的要求,因为Braker会尝试从另一个环境变量的位置猜测它们( $AUGUSTUS_CONFIG_PATH ),或者可以将两个目录作为命令行参数提供给braker.pl ,但我们建议我们建议将它们添加到您的$PATH变量中。对于您当前的狂欢会话,请输入:
PATH=:/your_path_to_augustus/bin/:/your_path_to_augustus/scripts/:$PATH
export PATH
对于所有bash会话,请在启动脚本(例如~/.bashrc )中添加上述行。
在ubuntu上,Python3通常默认安装,默认情况下, python3将在您的$PATH变量中,而Braker将自动找到它。但是,您可以选择以其他两种方式指定python3二进制位置:
导出环境变量$PYTHON3_PATH ,例如您的~/.bashrc文件中:
export PYTHON3_PATH=/path/to/python3/
指定命令行选项--PYTHON3_PATH=/path/to/python3/ to braker.pl 。
下载BAMTools(例如git clone https://github.com/pezmaster31/bamtools.git )。通过在外壳中键入以下内容来安装BAMTools:
cd your-bamtools-directory mkdir build cd build cmake .. make
如果已经在您的$PATH变量中,则Braker会自动找到BAMTools。否则,Braker可以使用环境变量$BAMTOOLS_PATH或使用命令行参数( --BAMTOOLS_PATH=/your_path_to_bamtools/bin/ f6 )来定位BAMTools二进制。为了设置环境变量,例如您当前的bash会话,请输入:
export BAMTOOLS_PATH=/your_path_to_bamtools/bin/
将上面的行添加到启动脚本(例如~/.bashrc ),以便为所有bash会话设置环境变量。
您可以使用NCBI BLAST+或钻石来去除冗余训练基因。您不需要这两个工具。如果存在钻石,它将是优选的,因为它要快得多。
获得并解开钻石,如下所示:
wget http://github.com/bbuchfink/diamond/releases/download/v0.9.24/diamond-linux64.tar.gz
tar xzf diamond-linux64.tar.gz
如果已经在您的$PATH变量中,则布拉克会自动找到钻石。否则,Braker可以使用环境变量$DIAMOND_PATH或使用命令行参数( --DIAMOND_PATH=/your_path_to_diamond )来定位钻石二进制。为了设置环境变量,例如您当前的bash会话,请输入:
export DIAMOND_PATH=/your_path_to_diamond/
将上面的行添加到启动脚本(例如~/.bashrc ),以便为所有bash会话设置环境变量。
如果您决定进行BLAST+,请使用sudo apt-get install ncbi-blast+安装NCBI BLAST+。
如果已经在您的$PATH变量中,则Braker会自动找到BlastP。否则,Braker可以使用环境变量$BLAST_PATH或使用命令行参数( --BLAST_PATH=/your_path_to_blast/ )来定位BLASTP二进制。为了设置环境变量,例如您当前的bash会话,请输入:
export BLAST_PATH=/your_path_to_blast/
将上面的行添加到启动脚本(例如~/.bashrc ),以便为所有bash会话设置环境变量。
Genemark-Etp需要以下工具,它将尝试将它们定位在您的$PATH变量中。因此,请确保将其位置添加到您的$PATH ,例如:
export PATH=$PATH:/your/path/to/Tool
对于下面的所有工具,将上面的行添加到启动脚本(例如~/.bashrc )中,以扩展所有bash会话的$PATH变量。
这些软件工具只有在您使用RNA-seq和蛋白质数据的Braker运行Braker之后才是强制性的!
stringTie2由genemark-etp使用来组装对齐的RNA-seq比对。可以从https://ccb.jhu.edu/software/stringtie/#install下载StringTie2的预编译版本。
如果您想同时使用RNA-seq和蛋白质数据运行Braker,则基因标准的软件包床托斯工具需要。您可以从https://github.com/arq5x/bedtools2/releases下载BedTools。在这里,您可以下载预编译版本的bedtools.static.binary ,例如
wget https://github.com/arq5x/bedtools2/releases/download/v2.30.0/bedtools.static.binary
mv bedtools.static.binary bedtools
chmod a+x
或者,您可以下载bedtools-2.30.0.tar.gz ,并使用make从源中进行编译,例如
wget https://github.com/arq5x/bedtools2/releases/download/v2.30.0/bedtools-2.30.0.tar.gz
tar -zxvf bedtools-2.30.0.tar.gz
cd bedtools2
make
有关更多信息,请参见https://bedtools.readthedocs.io/en/latest/content/installation.html。
GFFREAD是基因标准-ETP所需的实用软件。它可以从https://github.com/gpertea/gffread/releases/download/v0.12.7/gffread-0.12.7.linux_x86_64.tar.gz下载make
wget https://github.com/gpertea/gffread/releases/download/v0.12.7/gffread-0.12.7.Linux_x86_64.tar.gz
tar xzf gffread-0.12.7.Linux_x86_64.tar.gz
cd gffread-0.12.7.Linux_x86_64
make
如果您的所有文件均已格式化,则不需要samtools即可在没有Genemark-Etp的情况下运行Braker(即正确的sequences都应具有简短且唯一的fasta名称)。如果您不确定所有文件是否正确fomatter,则安装SamTool可能会有所帮助,因为Braker可以使用SamTool自动解决某些格式问题。
作为Samtools的先决条件,下载并安装htslib (例如git clone https://github.com/samtools/htslib.git ,请按照htslib文档进行安装)。
下载并安装samtools(例如git clone git://github.com/samtools/samtools.git ),随后遵循Samtools文档进行安装)。
如果已经在您的$PATH变量中,则Braker会自动找到Samtools。否则,Braker可以通过获取命令行参数( --SAMTOOLS_PATH=/your_path_to_samtools/ )或使用环境变量$SAMTOOLS_PATH来找到SamTools。对于导出变量,例如,对于当前的bash会话,请输入:
export SAMTOOLS_PATH=/your_path_to_samtools/
将上面的行添加到启动脚本(例如~/.bashrc ),以便为所有bash会话设置环境变量。
如果安装了Biopython,Braker可以通过Augustus预测的编码序列和蛋白质序列生成Fasta-Files,并生成轨道数据中心,以可视化使用MakeHub R16运行的Braker。这些是可选步骤。第一个可以使用命令行标志--skipGetAnnoFromFasta禁用,第二个可以通过使用命令行选项--makehub [email protected]来激活第二个,如果不需要这些可选步骤。应执行。
在Ubuntu上,安装Python3软件包管理器:
`sudo apt-get install python3-pip`
然后,使用以下方式安装Biopython
`sudo pip3 install biopython`
Braker要求CDBFASTA和CDBYANK使用Augustus脚本FIX_IN_FRAME_STOP_CODON_GENES.PY纠正使用框架终止密码子(拼接终止密码子)纠正奥古斯都基因。可以跳过--skip_fixing_broken_genes 。
在Ubuntu上,安装CDBFASTA:
sudo apt-get install cdbfasta
对于其他系统,您可以从https://github.com/gpertea/cdbfasta获得CDBFASTA,例如:
git clone https://github.com/gpertea/cdbfasta.git
cd cdbfasta
make all
在Ubuntu上,CDBFASTA和CDBYANK将在您的$PATH变量中安装,Braker将自动找到它们。但是,您可以选择以其他两种方式指定cdbfasta和cdbyank二进制位置:
$CDBTOOLS_PATH ,例如您的~/.bashrc文件中: export CDBTOOLS_PATH=/path/to/cdbtools/
--CDBTOOLS_PATH=/path/to/cdbtools/ to braker.pl 。 注意:对布拉克内部的独立spaln(原刺)的支持被贬低。
如果您运行原始启动,或者您想使用蛋白质与Braker一起使用蛋白质对齐,则需要此工具。只有在可以使用距离目标基因组的短进化距离的带注释的物种时,才能使用spaln在螺母之外。我们建议您通过Braker的Prothint运行Spaln。 Prothint带来了一个spaln二进制。如果您对系统不起作用,请从https://github.com/ogotoh/spaln下载Spaln。根据spaln/doc/SpalnReadMe22.pdf解开包装并安装。
Braker将尝试使用环境变量$ALIGNMENT_TOOL_PATH来定位SPALN可执行文件。另外,可以将其作为命令行参数( --ALIGNMENT_TOOL_PATH=/your/path/to/spaln )提供。
仅当您需要将UTR(从RNA-Seq数据)添加到预测基因或要训练Augustus的UTR参数并用UTR预测基因时,才需要此工具。无论如何,Gushr需要RNA-Seq数据的输入。
Gushr可在https://github.com/gaius-augustus/gushr上下载。通过键入获取它:
git clone https://github.com/Gaius-Augustus/GUSHR.git
Gushr执行Gemoma Jar文件R19, R20, R21 ,此JAR文件需要Java 1.8。在Ubuntu上,您可以使用以下命令安装Java 1.8:
sudo apt-get install openjdk-8-jdk
如果您的系统上安装了几个Java版本
sudo update-alternatives --config java
并选择正确的版本。
如果您切换--UTR=on ,bamtowig.py将需要以下可以从http://hgdownload.soe.ucsc.edu/admin/exe下载的工具:
Twobitinfo
fatotwobit
将这些工具安装到您的$路径中是可选的。如果您不这样做,然后切换--UTR=on ,bamtowig.py将它们自动下载到工作目录中。
如果您希望自动生成Braker Run的轨道数据中心,则需要在https://github.com/gaius-augustus/makehub上获得MakeHub软件。下载该软件(通过运行git clone https://github.com/Gaius-Augustus/MakeHub.git ,或从https://github.com/gaius-augustus/makehub/releases中获取版本软件包如果您下载了版本(例如unzip MakeHub.zip或tar -zxvf MakeHub.tar.gz 。
Braker将尝试使用环境变量$MAKEHUB_PATH来定位make_hub.py脚本。另外,可以将其作为命令行参数( --MAKEHUB_PATH=/your/path/to/MakeHub/ )提供。 Braker还可以尝试猜测系统上的MakeHub的位置。
如果您希望Braker从NCBI的SRA下载RNA-Seq库,则需要SRA工具包。您可以从http://daehwankimlab.github.io/hisat2/download/download/#version-hisat2-221获取SRA工具包的预编译版本。
Braker将尝试通过使用环境变量$SRATOOLS_PATH从SRA工具包(FastQ-Dump,Prefetch)中找到可执行的二进制文件。另外,可以将其作为命令行参数( --SRATOOLS_PATH=/your/path/to/SRAToolkit/ )提供。如果可执行文件在您的$PATH变量中,则布拉克还可以尝试猜测系统上SRA工具包的位置。
如果您想使用未对齐的RNA-Seq读取,则需要使用HISAT2软件将其映射到基因组中。可以从http://daehwankimlab.github.io/hisat2/download/download/#version-hisat2-221下载了预编译的HISAT2版本。
Braker将尝试通过使用环境变量$HISAT2_PATH来找到可执行的HISAT2二进制文件(HISAT2,HISAT2-build)。另外,可以将其作为命令行参数( --HISAT2_PATH=/your/path/to/HISAT2/ )提供。如果可执行文件位于您的$PATH变量中,则Braker还可以尝试猜测HISAT2在您的系统上的位置。
如果您想以Busco完整性模式在Braker中运行Tsebra,则需要安装REPLEASM。
wget https://github.com/huangnengCSU/compleasm/releases/download/v0.2.4/compleasm-0.2.4_x64-linux.tar.bz2
tar -xvjf compleasm-0.2.4_x64-linux.tar.bz2 &&
将结果文件夹creperm_kit添加到您的$PATH变量,例如:
export PATH=$PATH:/your/path/to/compleasm_kit
REPLEASM需要熊猫,可以安装以下方式:
pip install pandas
Braker(Braker.pl)使用GetConf查看系统上可以运行多少个线程。在Ubuntu上,您可以安装它:
sudo apt-get install libc-bin
在下文中,我们描述了“典型” Braker要求使用不同的输入数据类型。通常,我们建议您在基因组序列上运行Braker,这些序列已被软卸载以进行重复。 Braker只能应用于已软卸载以进行重复的基因组!
这种方法适用于具有良好转录组覆盖范围的RNA-seq库的物种基因组,并且蛋白质数据不在手头上。该管道如图2所示。
BRAKER has several ways to receive RNA-Seq data as input:
You can provide ID(s) of RNA-Seq libraries from SRA (in case of multiple IDs, separate them by comma) as argument to --rnaseq_sets_ids . The libraries belonging to the IDs are then downloaded automatically by BRAKER, eg:
braker.pl --species=yourSpecies --genome=genome.fasta
--rnaseq_sets_ids=SRA_ID1,SRA_ID2
You can use local FASTQ file(s) of unaligned reads as input. In this case, you have to provide BRAKER with the ID(s) of the RNA-Seq set(s) as argument to --rnaseq_sets_ids and the path(s) to the directories, where the FASTQ files are located as argument to --rnaseq_sets_dirs . For each ID ID , BRAKER will search in these directories for one FASTQ file named ID.fastq if the reads are unpaired, or for two FASTQ files named ID_1.fastq and ID_2.fastq if they are paired.
For example, if you have a paired library called 'SRA_ID1' and an unpaired library named 'SRA_ID2', you have to have a directory /path/to/local/fastq/files/ , where the files SRA_ID1_1.fastq , SRA_ID1_2.fastq , and SRA_ID2.fastq reside. Then, you could run BRAKER with following command:
braker.pl --species=yourSpecies --genome=genome.fasta
--rnaseq_sets_ids=SRA_ID1,SRA_ID2
--rnaseq_sets_dirs=/path/to/local/fastq/files/
There are two ways of supplying BRAKER with RNA-Seq data as bam file(s). First, you can do it in the same way as you would supply FASTQ file(s): Provide the ID(s)/name(s) of your bam file(s) as argument to --rnaseq_sets_ids and specify directories where the bam files reside with --rnaseq_sets_dirs . BRAKER will automatically detect that these ID(s) are bam and not FASTQ file(s), eg:
braker.pl --species=yourSpecies --genome=genome.fasta
--rnaseq_sets_ids=BAM_ID1,BAM_ID2
--rnaseq_sets_dirs=/path/to/local/bam/files/
Second, you can specify the paths to your bam file(s) directly, eg can either extract RNA-Seq spliced alignment information from bam files, or it can use such extracted information, directly.
braker.pl --species=yourSpecies --genome=genome.fasta
--bam=file1.bam,file2.bam
Please note that we generally assume that bam files were generated with HiSat2 because that is the aligner that would also be executed by BRAKER3 with fastq input. If you want for some reason to generate the bam files with STAR, use the option --outSAMstrandField intronMotif of STAR to produce files that are compatible wiht StringTie in BRAKER3.
In order to run BRAKER with RNA-Seq spliced alignment information that has already been extracted, run:
braker.pl --species=yourSpecies --genome=genome.fasta
--hints=hints1.gff,hints2.gff
The format of such a hints file must be as follows (tabulator separated file):
chrName b2h intron 6591 8003 1 + . pri=4;src=E
chrName b2h intron 6136 9084 11 + . mult=11;pri=4;src=E
...
The source b2h in the second column and the source tag src=E in the last column are essential for BRAKER to determine whether a hint has been generated from RNA-Seq data.
It is also possible to provide RNA-Seq sets in different ways for the same BRAKER run, any combination of above options is possible. It is not recommended to provide RNA-Seq data with --hints if you run BRAKER in ETPmode (RNA-Seq and protein data), because GeneMark-ETP won't use these hints!
This approach is suitable for genomes of species for which no RNA-Seq libraries are available. A large database of proteins (with possibly longer evolutionary distance to the target species) should be used in this case. This mode is illustrated in figure 9.
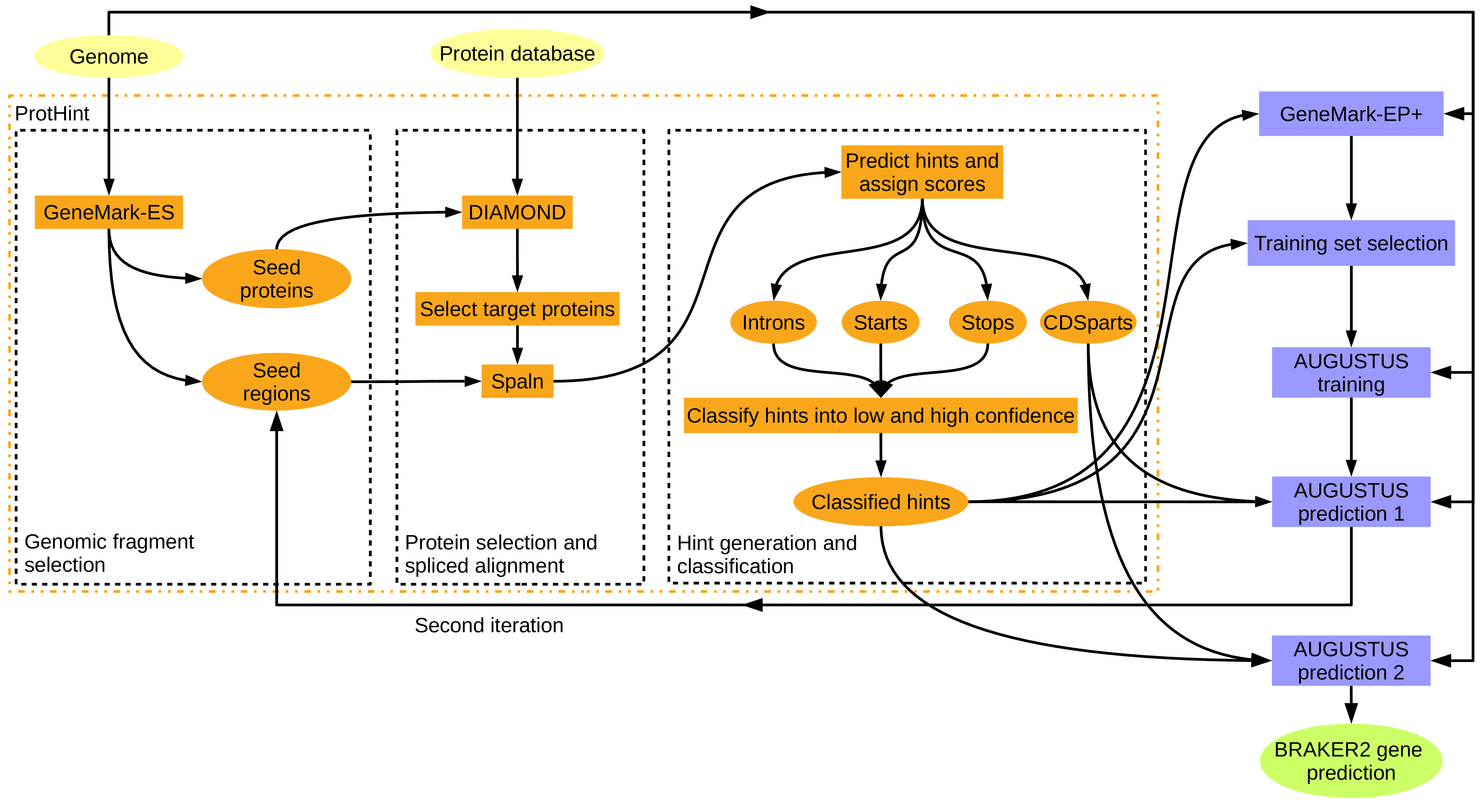
Figure 9: BRAKER with proteins of any evolutionary distance. ProtHint protein mapping pipelines is used to generate protein hints. ProtHint automatically determines which alignments are from close relatives, and which are from rather distant relatives.
For running BRAKER in this mode, type:
braker.pl --genome=genome.fa --prot_seq=proteins.fa
We recommend using OrthoDB as basis for proteins.fa . The instructions on how to prepare the input OrthoDB proteins are documented here: https://github.com/gatech-genemark/ProtHint#protein-database-preparation.
You can of course add additional protein sequences to that file, or try with a completely different database. Any database will need several representatives for each protein, though.
Instead of having BRAKER run ProtHint, you can also start BRAKER with hints already produced by ProtHint, by providing ProtHint's prothint_augustus.gff output:
braker.pl --genome=genome.fa --hints=prothint_augustus.gff
The format of prothint_augustus.gff in this mode looks like this:
2R ProtHint intron 11506230 11506648 4 + . src=M;mult=4;pri=4
2R ProtHint intron 9563406 9563473 1 + . grp=69004_0:001de1_702_g;src=C;pri=4;
2R ProtHint intron 8446312 8446371 1 + . grp=43151_0:001cae_473_g;src=C;pri=4;
2R ProtHint intron 8011796 8011865 2 - . src=P;mult=1;pri=4;al_score=0.12;
2R ProtHint start 234524 234526 1 + . src=P;mult=1;pri=4;al_score=0.08;
The prediction of all hints with src=M will be enforced. Hints with src=C are 'chained evidence', ie they will only be incorporated if all members of the group (grp=...) can be incorporated in a single transcript. All other hints have src=P in the last column. Supported features in column 3 are intron , start , stop and CDSpart .
If RNA-Seq (and only RNA-Seq) data is provided to BRAKER as a bam-file, and if the genome is softmasked for repeats, BRAKER can automatically train UTR parameters for AUGUSTUS. After successful training of UTR parameters, BRAKER will automatically predict genes including coverage information form RNA-Seq data. Example call:
braker.pl --species=yourSpecies --genome=genome.fasta
--bam=file.bam --UTR=on
警告:
This feature is experimental!
--UTR=on is currently not compatible with bamToWig.py as released in AUGUSTUS 3.3.3; it requires the current development code version from the github repository (git clone https://github.com/Gaius-Augustus/Augustus.git).
--UTR=on increases memory consumption of AUGUSTUS. Carefully monitor jobs if your machine was close to maxing RAM without --UTR=on! Reducing the number of cores will also reduce RAM consumption.
UTR prediction sometimes improves coding sequence prediction accuracy, but not always. If you try this feature, carefully compare results with and without UTR parameters, afterwards (eg in UCSC Genome Browser).
For running BRAKER without UTR parameters, it is not very important whether RNA-Seq data was generated by a stranded protocol (because spliced alignments are 'artificially stranded' by checking the splice site pattern). However, for UTR training and prediction, stranded libraries may provide information that is valuable for BRAKER.
After alignment of the stranded RNA-Seq libraries, separate the resulting bam file entries into two files: one for plus strand mappings, one for minus strand mappings. Call BRAKER as follows:
braker.pl --species=yourSpecies --genome=genome.fasta
--bam=plus.bam,minus.bam --stranded=+,-
--UTR=on
You may additionally include bam files from unstranded libraries. Those files will not used for generating UTR training examples, but they will be included in the final gene prediction step as unstranded coverage information, example call:
braker.pl --species=yourSpecies --genome=genome.fasta
--bam=plus.bam,minus.bam,unstranded.bam
--stranded=+,-,. --UTR=on
Warning: This feature is experimental and currently has low priority on our maintenance list!
The native mode for running BRAKER with RNA-Seq and protein data. This will call GeneMark-ETP, which will use RNA-Seq and protein hints for training GeneMark-ETP. Subsequently, AUGUSTUS is trained on 'high-confindent' genes (genes with very high extrinsic evidence support) from the GeneMark-ETP prediction and a set of genes is predicted by AUGUSTUS. In a last step, the predictions of AUGUSTUS and GeneMark-ETP are combined using TSEBRA.
Alignment of RNA-Seq reads
GeneMark-ETP utilizes Stringtie2 to assemble RNA-Seq data, which requires that the aligned reads (BAM files) contain the XS (strand) tag for spliced reads. Therefore, if you align your reads with HISAT2, you must enable the --dta option, or if you use STAR, you must use the --outSAMstrandField intronMotif option. TopHat alignments include this tag by default.
To call the pipeline in this mode, you have to provide it with a protein database using --prot_seq (as described in BRAKER with protein data), and RNA-Seq data either by their SRA ID so that they are downloaded by BRAKER, as unaligned reads in FASTQ format, and/or as aligned reads in bam format (as described in BRAKER with RNA-Seq data). You could also specify already processed extrinsic evidence using the --hints option. However, this is not recommend for a normal BRAKER run in ETPmode, as these hints won't be used in the GeneMark-ETP step. Only use --hints when you want to skip the GenMark-ETP step!
Examples of how you could run BRAKER in ETPmode:
braker.pl --genome=genome.fa --prot_seq=orthodb.fa
--rnaseq_sets_ids=SRA_ID1,SRA_ID2
--rnaseq_sets_dirs=/path/to/local/RNA-Seq/files/
braker.pl --genome=genome.fa --prot_seq=orthodb.fa
--rnaseq_sets_ids=SRA_ID1,SRA_ID2,SRA_ID3
braker.pl --genome=genome.fa --prot_seq=orthodb.fa
--bam=/path/to/SRA_ID1.bam,/path/to/SRA_ID2.bam
A preliminary protocol for integration of assembled subreads from PacBio ccs sequencing in combination with short read Illumina RNA-Seq and protein database is described at https://github.com/Gaius-Augustus/BRAKER/blob/master/docs/long_reads/long_read_protocol .md
We forked GeneMark-ETP and hard coded that StringTie will perform long read assembly in that particular version. If you want to use this 'fast-hack' version for BRAKER, you have to prepare the BAM file with long read to genome spliced alignments outside of BRAKER, eg:
T=48 # adapt to your number of threads
minimap2 -t${T} -ax splice:hq -uf genome.fa isoseq.fa > isoseq.sam
samtools view -bS --threads ${T} isoseq.sam -o isoseq.bam
Pull the adapted container:
singularity build braker3_lr.sif docker://teambraker/braker3:isoseq
Calling BRAKER3 with a BAM file of spliced-aligned IsoSeq Reads:
singularity exec -B ${PWD}:${PWD} braker3_lr.sif braker.pl --genome=genome.fa --prot_seq=protein_db.fa –-bam=isoseq.bam --threads=${T}
Warning Do NOT mix short read and long read data in this BRAKER/GeneMark-ETP variant!
Warning The accuracy of gene prediction here heavily depends on the depth of your isoseq data. We verified with PacBio HiFi reads from 2022 that given sufficient completeness of the assembled transcriptome you will reach similar results as with short reads. However, we also observed a drop in accuracy compared to short reads when using other long read data sets with higher error rates and less sequencing depth.
Please run braker.pl --help to obtain a full list of options.
Compute AUGUSTUS ab initio predictions in addition to AUGUSTUS predictions with hints (additional output files: augustus.ab_initio.* . This may be useful for estimating the quality of training gene parameters when inspecting predictions in a Browser.
One or several command line arguments to be passed to AUGUSTUS, if several arguments are given, separate them by whitespace, ie "--first_arg=sth --second_arg=sth" . This may be be useful if you know that gene prediction in your particular species benefits from a particular AUGUSTUS argument during the prediction step.
Specifies the maximum number of threads that can be used during computation. BRAKER has to run some steps on a single thread, others can take advantage of multiple threads. If you use more than 8 threads, this will not speed up all parallelized steps, in particular, the time consuming optimize_augustus.pl will not use more than 8 threads. However, if you don't mind some threads being idle, using more than 8 threads will speed up other steps.
GeneMark-ETP option: run algorithm with branch point model. Use this option if you genome is a fungus.
Use the present config and parameter files if they exist for 'species'; will overwrite original parameters if BRAKER performs an AUGUSTUS training.
Execute CRF training for AUGUSTUS; resulting parameters are only kept for final predictions if they show higher accuracy than HMM parameters. This increases runtime!
Change the parameter
Generate UTR training examples for AUGUSTUS from RNA-Seq coverage information, train AUGUSTUS UTR parameters and predict genes with AUGUSTUS and UTRs, including coverage information for RNA-Seq as evidence. This is an experimental feature!
If you performed a BRAKER run without --UTR=on, you can add UTR parameter training and gene prediction with UTR parameters (and only RNA-Seq hints) with the following command:
braker.pl --genome=../genome.fa --addUTR=on
--bam=../RNAseq.bam --workingdir=$wd
--AUGUSTUS_hints_preds=augustus.hints.gtf
--threads=8 --skipAllTraining --species=somespecies
Modify augustus.hints.gtf to point to the AUGUSTUS predictions with hints from previous BRAKER run; modify flaning_DNA value to the flanking region from the log file of your previous BRAKER run; modify some_new_working_directory to the location where BRAKER should store results of the additional BRAKER run; modify somespecies to the species name used in your previous BRAKER run.
Add UTRs from RNA-Seq converage information to AUGUSTUS gene predictions using GUSHR. No training of UTR parameters and no gene prediction with UTR parameters is performed.
If you performed a BRAKER run without --addUTR=on, you can add UTRs results of a previous BRAKER run with the following command:
braker.pl --genome=../genome.fa --addUTR=on
--bam=../RNAseq.bam --workingdir=$wd
--AUGUSTUS_hints_preds=augustus.hints.gtf --threads=8
--skipAllTraining --species=somespecies
Modify augustus.hints.gtf to point to the AUGUSTUS predictions with hints from previous BRAKER run; modify some_new_working_directory to the location where BRAKER should store results of the additional BRAKER run; this run will not modify AUGUSTUS parameters. We recommend that you specify the original species of the original run with --species=somespecies . Otherwise, BRAKER will create an unneeded species parameters directory Sp_* .
If --UTR=on is enabled, strand-separated bam-files can be provided with --bam=plus.bam,minus.bam . In that case, --stranded=... should hold the strands of the bam files ( + for plus strand, - for minus strand, . for unstranded). Note that unstranded data will be used in the gene prediction step, only, if the parameter --stranded=... is set. This is an experimental feature! GUSHR currently does not take advantage of stranded data.
If --makehub and [email protected] (with your valid e-mail adress) are provided, a track data hub for visualizing results with the UCSC Genome Browser will be generated using MakeHub (https://github.com/Gaius-Augustus/MakeHub).
By default, GeneMark-ES/ET/EP/ETP uses a probability of 0.001 for predicting the donor splice site pattern GC (instead of GT). It may make sense to increase this value for species where this donor splice site is more common. For example, in the species Emiliania huxleyi , about 50% of donor splice sites have the pattern GC (https://media.nature.com/original/nature-assets/nature/journal/v499/n7457/extref/nature12221-s2.pdf, page 5).
Use a species-specific lineage, eg arthropoda_odb10 for an arthropod. BRAKER does not support auto-typing of the lineage.
Specifying a BUSCO-lineage invokes two changes in BRAKER R28 :
BRAKER will run compleasm with the specified lineage in genome mode and convert the detected BUSCO matches into hints for AUGUSTUS. This may increase the number of BUSCOs in the augustus.hints.gtf file slightly.
BRAKER will invoke best_by_compleasm.py to check whether the braker.gtf file that is by default generated by TSEBRA has the lowest amount of missing BUSCOs compared to the augustus.hints.gtf and the genemark.gtf file. If not, the following decision schema is applied to re-run TSEBRA to minimize the missing BUSCOs in the final output of BRAKER (always braker.gtf). If an alternative and better gene set is created, the original braker.gtf gene set is moved to a directory called braker_original. Information on what happened during the best_by_compleasm.py run is written to the file best_by_compleasm.log.
![best_by_busco[fig14]](https://images.downcodes.com/uploads/20250214/img_67aee79a11fd439.png)
Please note that using BUSCO to assess the quality of a gene set, in particular when comparing BRAKER to other pipelines, does not make sense once you specified a BUSCO lineage. We recommend that you use other measures to assess the quality of your gene set, eg by comparing it to a reference gene set or running OMArk.
BRAKER produces several important output files in the working directory.
braker.gtf: Final gene set of BRAKER. This file may contain different contents depending on how you called BRAKER
in ETPmode: Final gene set of BRAKER consisting of genes predicted by AUGUSTUS and GeneMark-ETP that were combined and filtered by TSEBRA.
otherwise: Union of augustus.hints.gtf and reliable GeneMark-ES/ET/EP predictions (genes fully supported by external evidence). In --esmode , this is the union of augustus.ab_initio.gtf and all GeneMark-ES genes. Thus, this set is generally more sensitive (more genes correctly predicted) and can be less specific (more false-positive predictions can be present). This output is not necessarily better than augustus.hints.gtf, and it is not recommended to use it if BRAKER was run in ESmode.
braker.codingseq: Final gene set with coding sequences in FASTA format
braker.aa: Final gene set with protein sequences in FASTA format
braker.gff3: Final gene set in gff3 format (only produced if the flag --gff3 was specified to BRAKER.
Augustus/*: Augustus gene set(s) in as gtf/conding/aa files
GeneMark-E*/genemark.gtf: Genes predicted by GeneMark-ES/ET/EP/EP+/ETP in GTF-format.
hintsfile.gff: The extrinsic evidence data extracted from RNAseq.bam and/or protein data.
braker_original/*: Genes predicted by BRAKER (TSEBRA merge) before compleasm was used to improve BUSCO completeness
bbc/*: output folder of best_by_compleasm.py script from TSEBRA that is used to improve BUSCO completeness in the final output of BRAKER
Output files may be present with the following name endings and formats:
Coding sequences in FASTA-format are produced if the flag --skipGetAnnoFromFasta was not set.
Protein sequence files in FASTA-format are produced if the flag --skipGetAnnoFromFasta was not set.
For details about gtf format, see http://www.sanger.ac.uk/Software/formats/GFF/. A GTF-format file contains one line per predicted exon.例子:
HS04636 AUGUSTUS initial 966 1017 . + 0 transcript_id "g1.1"; gene_id "g1";
HS04636 AUGUSTUS internal 1818 1934 . + 2 transcript_id "g1.1"; gene_id "g1";
The columns (fields) contain:
seqname source feature start end score strand frame transcript ID and gene ID
If the --makehub option was used and MakeHub is available on your system, a hub directory beginning with the name hub_ will be created. Copy this directory to a publicly accessible web server. A file hub.txt resides in the directory. Provide the link to that file to the UCSC Genome Browser for visualizing results.
An incomplete example data set is contained in the directory BRAKER/example . In order to complete the data set, please download the RNA-Seq alignment file (134 MB) with wget :
cd BRAKER/example
wget http://topaz.gatech.edu/GeneMark/Braker/RNAseq.bam
In case you have trouble accessing that file, there's also a copy available from another server:
cd BRAKER/example
wget http://bioinf.uni-greifswald.de/augustus/datasets/RNAseq.bam
The example data set was not compiled in order to achieve optimal prediction accuracy, but in order to quickly test pipeline components. The small subset of the genome used in these test examples is not long enough for BRAKER training to work well.
Data corresponds to the last 1,000,000 nucleotides of Arabidopsis thaliana 's chromosome Chr5, split into 8 artificial contigs.
RNA-Seq alignments were obtained by VARUS.
The protein sequences are a subset of OrthoDB v10 plants proteins.
List of files:
genome.fa - genome file in fasta formatRNAseq.bam - RNA-Seq alignment file in bam format (this file is not a part of this repository, it must be downloaded separately from http://topaz.gatech.edu/GeneMark/Braker/RNAseq.bam)RNAseq.hints - RNA-Seq hints (can be used instead of RNAseq.bam as RNA-Seq input to BRAKER)proteins.fa - protein sequences in fasta formatThe below given commands assume that you configured all paths to tools by exporting bash variables or that you have the necessary tools in your $PATH.
The example data set also contains scripts tests/test*.sh that will execute below listed commands for testing BRAKER with the example data set. You find example results of AUGUSTUS and GeneMark-ES/ET/EP/ETP in the folder results/test* . Be aware that BRAKER contains several parts where random variables are used, ie results that you obtain when running the tests may not be exactly identical. To compare your test results with the reference ones, you can use the compare_intervals_exact.pl script as follows:
# Compare CDS features
compare_intervals_exact.pl --f1 augustus.hints.gtf --f2 ../../results/test${N}/augustus.hints.gtf --verbose
# Compare transcripts
compare_intervals_exact.pl --f1 augustus.hints.gtf --f2 ../../results/test${N}/augustus.hints.gtf --trans --verbose
Several tests use --gm_max_intergenic 10000 option to make the test runs faster. It is not recommended to use this option in real BRAKER runs, the speed increase achieved by adjusting this option is negligible on full-sized genomes.
We give runtime estimations derived from computing on Intel(R) Xeon(R) CPU E5530 @ 2.40GHz .
The following command will run the pipeline according to Figure 3:
braker.pl --genome genome.fa --bam RNAseq.bam --threads N --busco_lineage=lineage_odb10
This test is implemented in test1.sh , expected runtime is ~20 minutes.
The following command will run the pipeline according to Figure 4:
braker.pl --genome genome.fa --prot_seq proteins.fa --threads N --busco_lineage=lineage_odb10
This test is implemented in test2.sh , expected runtime is ~20 minutes.
The following command will run a pipeline that first trains GeneMark-ETP with protein and RNA-Seq hints and subsequently trains AUGUSTUS on the basis of GeneMark-ETP predictions. AUGUSTUS predictions are also performed with hints from both sources, see Figure 5.
Run with local RNA-Seq file:
braker.pl --genome genome.fa --prot_seq proteins.fa --bam ../RNAseq.bam --threads N --busco_lineage=lineage_odb10
This test is implemented in test3.sh , expected runtime is ~20 minutes.
Download RNA-Seq library from Sequence Read Archive (~1gb):
braker.pl --genome genome.fa --prot_seq proteins.fa --rnaseq_sets_ids ERR5767212 --threads N --busco_lineage=lineage_odb10
This test is implemented in test3_4.sh , expected runtime is ~35 minutes.
The training step of all pipelines can be skipped with the option --skipAllTraining . This means, only AUGUSTUS predictions will be performed, using pre-trained, already existing parameters. For example, you can predict genes with the command:
braker.pl --genome=genome.fa --bam RNAseq.bam --species=arabidopsis
--skipAllTraining --threads N
This test is implemented in test4.sh , expected runtime is ~1 minute.
The following command will run the pipeline with no extrinsic evidence:
braker.pl --genome=genome.fa --esmode --threads N
This test is implemented in test5.sh , expected runtime is ~20 minutes.
The following command will run BRAKER with training UTR parameters from RNA-Seq coverage data:
braker.pl --genome genome.fa --bam RNAseq.bam --UTR=on --threads N
This test is implemented in test6.sh , expected runtime is ~20 minutes.
The following command will add UTRs to augustus.hints.gtf from RNA-Seq coverage data:
braker.pl --genome genome.fa --bam RNAseq.bam --addUTR=on --threads N
This test is implemented in test7.sh , expected runtime is ~20 minutes.
There is currently no clean way to restart a failed BRAKER run (after solving some problem). However, it is possible to start a new BRAKER run based on results from a previous run -- given that the old run produced the required intermediate results. We will in the following refer to the old working directory with variable ${BRAKER_OLD} , and to the new BRAKER working directory with ${BRAKER_NEW} . The file what-to-cite.txt will always only refer to the software that was actually called by a particular run. You might have to combine the contents of ${BRAKER_NEW}/what-to-cite.txt with ${BRAKER_OLD}/what-to-cite.txt for preparing a publication. The following figure illustrates at which points BRAKER run may be intercepted.
![braker-intercept[fig8]](https://images.downcodes.com/uploads/20250214/img_67aee79a12cab310.png)
Figure 10: Points for intercepting a BRAKER run and reusing intermediate results in a new BRAKER run.
This option is only possible for BRAKER in ETmode or EPmode and不是in ETPmode!
If you have access to an existing BRAKER output that contains hintsfiles that were generated from extrinsic data, such as RNA-Seq or protein sequences, you can recycle these hints files in a new BRAKER run. Also, hints from a separate ProtHint run can be directly used in BRAKER.
The hints can be given to BRAKER with --hints ${BRAKER_OLD}/hintsfile.gff option. This is illustrated in the test files test1_restart1.sh , test2_restart1.sh , test4_restart1.sh . The other modes (for which this test is missing) cannot be restarted in this way.
The GeneMark result can be given to BRAKER with --geneMarkGtf ${BRAKER_OLD}/GeneMark*/genemark.gtf option if BRAKER is run in ETmode or EPmode. This is illustrated in the test files test1_restart2.sh , test2_restart2.sh , test5_restart2.sh .
In ETPmode, you can either provide BRAKER with the results of the GeneMarkETP step manually, with --geneMarkGtf ${BRAKER_OLD}/GeneMark-ETP/proteins.fa/genemark.gtf , --traingenes ${BRAKER_OLD}/GeneMark-ETP/training.gtf , and --hints ${BRAKER_OLD}/hintsfile.gff (see test3_restart1.sh for an example), or you can specify the previous GeneMark-ETP results with the option --gmetp_results_dir ${BRAKER_OLD}/GeneMark-ETP/ so that BRAKER can search for the files automatically (see test3_restart2.sh for an example).
The trained species parameters for AGUSTUS can be passed with --skipAllTraining and --species $speciesName options. This is illustrated in test*_restart3.sh files. Note that in ETPmode you have to specify the GeneMark files as described in Option 2!
Before reporting bugs, please check that you are using the most recent versions of GeneMark-ES/ET/EP/ETP, AUGUSTUS and BRAKER. Also, check the list of Common problems, and the Issue list on GitHub before reporting bugs. We do monitor open issues on GitHub. Sometimes, we are unable to help you, immediately, but we try hard to solve your problems.
If you found a bug, please open an issue at https://github.com/Gaius-Augustus/BRAKER/issues (or contact [email protected] or [email protected]).
Information worth mentioning in your bug report:
Check in braker/yourSpecies/braker.log at which step braker.pl crashed.
There are a number of other files that might be of interest, depending on where in the pipeline the problem occurred. Some of the following files will not be present if they did not contain any errors.
braker/yourSpecies/errors/bam2hints.*.stderr - will give details on a bam2hints crash (step for converting bam file to intron gff file)
braker/yourSpecies/hintsfile.gff - is this file empty? If yes, something went wrong during hints generation - does this file contain hints from source “b2h” and of type “intron”? If not: GeneMark-ET will not be able to execute properly. Conversely, GeneMark-EP+ will not be able to execute correctly if hints from the source "ProtHint" are missing.
braker/yourSpecies/spaln/*err - errors reported by spaln
braker/yourSpecies/errors/GeneMark-{ET,EP,ETP}.stderr - errors reported by GeneMark-ET/EP+/ETP
braker/yourSpecies/errors/GeneMark-{ET,EP,ETP).stdout - may give clues about the point at which errors in GeneMark-ET/EP+/ETP occured
braker/yourSpecies/GeneMark-{ET,EP,ETP}/genemark.gtf - is this file empty? If yes, something went wrong during executing GeneMark-ET/EP+/ETP
braker/yourSpecies/GeneMark-{ET,EP}/genemark.f.good.gtf - is this file empty? If yes, something went wrong during filtering GeneMark-ET/EP+ genes for training AUGUSTUS
braker/yourSpecies/genbank.good.gb - try a “grep -c LOCUS genbank.good.gb” to determine the number of training genes for training AUGUSTUS, should not be low
braker/yourSpecies/errors/firstetraining.stderr - contains errors from first iteration of training AUGUSTUS
braker/yourSpecies/errors/secondetraining.stderr - contains errors from second iteration of training AUGUSTUS
braker/yourSpecies/errors/optimize_augustus.stderr - contains errors optimize_augustus.pl (additional training set for AUGUSTUS)
braker/yourSpecies/errors/augustus*.stderr - contain AUGUSTUS execution errors
braker/yourSpecies/startAlign.stderr - if you provided a protein fasta file, something went wrong during protein alignment
braker/yourSpecies/startAlign.stdout - may give clues on at which point protein alignment went wrong
BRAKER complains that the RNA-Seq file does not correspond to the provided genome file, but I am sure the files correspond to each other!
Please check the headers of the genome FASTA file. If the headers are long and contain whitespaces, some RNA-Seq alignment tools will truncate sequence names in the BAM file. This leads to an error with BRAKER. Solution: shorten/simplify FASTA headers in the genome file before running the RNA-Seq alignment and BRAKER.
GeneMark fails!
(a) GeneMark by default only uses contigs longer than 50k for training. If you have a highly fragmented assembly, this might lead to "no data" for training. You can override the default minimal length by setting the BRAKER argument --min_contig=10000 .
(b) see "[something] failed to execute" below.
[something] failed to execute!
When providing paths to software to BRAKER, please use absolute, non-abbreviated paths. For example, BRAKER might have problems with --SAMTOOLS_PATH=./samtools/ or --SAMTOOLS_PATH=~/samtools/ . Please use SAMTOOLS_PATH=/full/absolute/path/to/samtools/ , instead. This applies to all path specifications as command line options to braker.pl . Relative paths and absolute paths will not pose problems if you export a bash variable, instead, or if you append the location of tools to your $PATH variable.
GeneMark-ETP in BRAKER dies with '/scratch/11232323': No such file or directory.
This appears to be related to sorting large files, and it's a system configuration depending problem. Solve it with export TMPDIR=/tmp/ before calling BRAKER via Singularity.
BRAKER cannot find the Augustus script XYZ...
Update Augustus from github with git clone https://github.com/Gaius-Augustus/Augustus.git . Do not use Augustus from other sources. BRAKER is highly dependent on an up-to-date Augustus. Augustus releases happen rather rarely, updates to the Augustus scripts folder occur rather frequently.
Does BRAKER depend on Python3?
确实如此。 The python scripts employed by BRAKER are not compatible with Python2.
Why does BRAKER predict more genes than I expected?
If transposable elements (or similar) have not been masked appropriately, AUGUSTUS tends to predict those elements as protein coding genes. This can lead to a huge number genes. You can check whether this is the case for your project by BLASTing (or DIAMONDing) the predicted protein sequences against themselves (all vs. all) and counting how many of the proteins have a high number of high quality matches. You can use the output of this analysis to divide your gene set into two groups: the protein coding genes that you want to find and the repetitive elements that were additionally predicted.
I am running BRAKER in Anaconda and something fails...
Update AUGUSTUS and BRAKER from github with git clone https://github.com/Gaius-Augustus/Augustus.git and git clone https://github.com/Gaius-Augustus/BRAKER.git . The Anaconda installation is great, but it relies on releases of AUGUSTUS and BRAKER - which are often lagging behind. Please use the current GitHub code, instead.
Why and where is the GenomeThreader support gone?
BRAKER is a joint project between teams from University of Greifswald and Georgia Tech. While the group of Mark Bordovsky from Georgia Tech contributes GeneMark expertise, the group of Mario Stanke from University of Greifswald contributes AUGUSTUS expertise. Using GenomeThreader to build training genes for AUGUSTUS in BRAKER circumvents execution of GeneMark. Thus, the GenomeThreader mode is strictly speaking not part of the BRAKER project. The previous functionality of BRAKER with GenomeThreader has been moved to GALBA at https://github.com/Gaius-Augustus/GALBA. Note that GALBA has also undergone extension for using Miniprot instead of GenomeThreader.
My BRAKER gene set has too many BUSCO duplicates!
AUGUSTUS within BRAKER can predict alternative splicing isoforms. Also the merge of the AUGUSTUS and GeneMark gene set by TSEBRA within BRAKER may result in additional isoforms for a single gene. The BUSCO duplicates usually come from alternative splicing isoforms, ie they are expected.
Augustus and/or etraining within BRAKER complain that the file aug_cmdln_parameters.json is missing. Even though I am using the latest Singularity container!
BRAKER copies the AUGUSTUS_CONFIG_PATH folder to a writable location. In older versions of Augustus, that file was indeed not existing. If the local writable copy of a folder already exists, BRAKER will not re-copy it. Simply delete the old folder. (It is often ~/.augustus , so you can simply do rm -rf ~/.augustus ; the folder might be residing in $PWD if your home directory was not writable).
I sit behind a firewall, compleasm cannot download the BUSCO files, what can I do? See Issue #785 (comment)
Since BRAKER is a pipeline that calls several Bioinformatics tools, publication of results obtained by BRAKER requires that not only BRAKER is cited, but also the tools that are called by BRAKER. BRAKER will output a file what-to-cite.txt in the BRAKER working directory, informing you about which exact sources apply to your run.
Always cite:
Stanke, M., Diekhans, M., Baertsch, R. and Haussler, D. (2008).使用天然和同步映射的cDNA比对来改善从头基因的发现。 Bioinformatics, doi: 10.1093/bioinformatics/btn013.
Stanke. M., Schöffmann, O., Morgenstern, B. and Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62.
If you provided any kind of evidence for BRAKER, cite:
If you provided both short read RNA-Seq evidence and a large database of proteins, cite:
Gabriel, L., Bruna, T., Hoff, KJ, Ebel, M., Lomsadze, A., Borodovsky, M., Stanke, M. (2023). BRAKER3: Fully Automated Genome Annotation Using RNA-Seq and Protein Evidence with GeneMark-ETP, AUGUSTUS and TSEBRA. bioRxiV, doi: 10.1101/2023.06.10.54444910.1101/2023.01.01.474747.
Bruna, T., Lomsadze, A., Borodovsky, M. (2023). GeneMark-ETP: Automatic Gene Finding in Eukaryotic Genomes in Consistence with Extrinsic Data. bioRxiv, doi: 10.1101/2023.01.13.524024.
Kovaka, S., Zimin, AV, Pertea, GM, Razaghi, R., Salzberg, SL, & Pertea, M. (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome biology, 20(1):1-13.
Pertea, G., & Pertea, M. (2020). GFF utilities: GffRead and GffCompare. F1000Research, 9.
Quinlan, AR (2014). BEDTools: the Swiss‐army tool for genome feature analysis. Current protocols in bioinformatics, 47(1):11-12.
If the only source of evidence for BRAKER was a large database of protein sequences, cite:
If the only source of evidence for BRAKER was RNA-Seq data, cite:
Hoff, KJ, Lange, S., Lomsadze, A., Borodovsky, M. and Stanke, M. (2016). BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics, 32(5):767-769.
Lomsadze, A., Paul DB, and Mark B. (2014) Integration of Mapped Rna-Seq Reads into Automatic Training of Eukaryotic Gene Finding Algorithm. Nucleic Acids Research 42(15): e119--e119
If you called BRAKER3 with an IsoSeq BAM file, or if you envoked the --busco_lineage option, cite:
If you called BRAKER with the --busco_lineage option, in addition, cite:
Simão, FA, Waterhouse, RM, Ioannidis, P., Kriventseva, EV, & Zdobnov, EM (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31(19), 3210-3212.
Li,H。(2023)。 Protein-to-genome alignment with miniprot. Bioinformatics, 39(1), btad014.
Huang, N., & Li, H. (2023). compleasm: a faster and more accurate reimplementation of BUSCO. Bioinformatics, 39(10), btad595.
If any kind of AUGUSTUS training was performed by BRAKER, check carefully whether you configured BRAKER to use NCBI BLAST or DIAMOND. One of them was used to filter out redundant training gene structures.
If you used NCBI BLAST, please cite:
Altschul, AF, Gish, W., Miller, W., Myers, EW and Lipman, DJ (1990). A basic local alignment search tool. J Mol Biol 215:403--410.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., and Madden, TL (2009). Blast+: architecture and applications. BMC bioinformatics, 10(1):421.
If you used DIAMOND, please cite:
If BRAKER was executed with a genome file and no extrinsic evidence, cite, then GeneMark-ES was used, cite:
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, YO and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Research, 33(20):6494--6506.
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, YO and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome research, pages gr--081612, 2008.
Hoff, KJ, Lomsadze, A., Borodovsky, M. and Stanke, M. (2019). Whole-Genome Annotation with BRAKER.方法摩尔生物。 1962:65-95, doi: 10.1007/978-1-4939-9173-0_5.
If BRAKER was run with proteins as source of evidence, please cite all tools that are used by the ProtHint pipeline to generate hints:
Bruna, T., Lomsadze, A., & Borodovsky, M. (2020). GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics and Bioinformatics, 2(2), lqaa026.
Buchfink, B., Xie, C., Huson, DH (2015).使用钻石快速和敏感的蛋白质对齐。 Nature Methods 12:59-60.
Lomsadze, A., Ter-Hovhannisyan, V., Chernoff, YO and Borodovsky, M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Research, 33(20):6494--6506.
Iwata, H., and Gotoh, O. (2012). Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic acids research, 40(20), e161-e161.
Gotoh, O., Morita, M., Nelson, DR (2014). Assessment and refinement of eukaryotic gene structure prediction with gene-structure-aware multiple protein sequence alignment. BMC bioinformatics, 15(1), 189.
If BRAKER was executed with RNA-Seq alignments in bam-format, then SAMtools was used, cite:
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R.; 1000 Genome Project Data Processing Subgroup (2009).序列比对/地图格式和Samtools。 Bioinformatics, 25(16):2078-9.
Barnett, DW, Garrison, EK, Quinlan, AR, Strömberg, MP and Marth GT (2011). BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics, 27(12):1691-2
If BRAKER downloaded RNA-Seq libraries from SRA using their IDs, cite SRA, SRA toolkit, and HISAT2:
Leinonen, R., Sugawara, H., Shumway, M., & International Nucleotide Sequence Database Collaboration. (2010)。 The sequence read archive. Nucleic acids research, 39(suppl_1), D19-D21.
SRA Toolkit Development Team (2020). SRA Toolkit. https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software.
Kim, D., Paggi, JM, Park, C., Bennett, C., & Salzberg, SL (2019).基于图的基因组对齐和基因分型与HISAT2和HISAT基因型。 Nature biotechnology, 37(8):907-915.
If BRAKER was executed using RNA-Seq data in FASTQ format, cite HISAT2:
If BRAKER called MakeHub for creating a track data hub for visualization of BRAKER results with the UCSC Genome Browser, cite:
If BRAKER called GUSHR for generating UTRs, cite:
Keilwagen, J., Hartung, F., Grau, J. (2019) GeMoMa: Homology-based gene prediction utilizing intron position conservation and RNA-seq data.方法摩尔生物。 1962:161-177, doi: 10.1007/978-1-4939-9173-0_9.
Keilwagen, J., Wenk, M., Erickson, JL, Schattat, MH, Grau, J., Hartung F. (2016) Using intron position conservation for homology-based gene prediction. Nucleic Acids Research, 44(9):e89.
Keilwagen, J., Hartung, F., Paulini, M., Twardziok, SO, Grau, J. (2018) Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics, 19(1):189.
All source code, ie scripts/*.pl or scripts/*.py are under the Artistic License (see http://www.opensource.org/licenses/artistic-license.php).
[F1] EX = ES/ET/EP/ETP, all available for download under the name GeneMark-ES/ET/EP ↩
[F2] Please use the latest version from the master branch of AUGUSTUS distributed by the original developers, it is available from github at https://github.com/Gaius-Augustus/Augustus. Problems have been reported from users that tried to run BRAKER with AUGUSTUS releases maintained by third parties, ie Bioconda. ↩
[F4] install with sudo apt-get install cpanminus ↩
[F6] The binary may eg reside in bamtools/build/src/toolkit ↩
[R0] Bruna, Tomas, Hoff, Katharina J., Lomsadze, Alexandre, Stanke, Mario, and Borodovsky, Mark. 2021. “BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database." NAR Genomics and Bioinformatics 3(1):lqaa108.↩
[R1] Hoff, Katharina J, Simone Lange, Alexandre Lomsadze, Mark Borodovsky, and Mario Stanke. 2015. “BRAKER1: Unsupervised Rna-Seq-Based Genome Annotation with Genemark-et and Augustus.” Bioinformatics 32 (5). Oxford University Press: 767--69.↩
[R2] Lomsadze, Alexandre, Paul D Burns, and Mark Borodovsky. 2014. “Integration of Mapped Rna-Seq Reads into Automatic Training of Eukaryotic Gene Finding Algorithm.” Nucleic Acids Research 42 (15). Oxford University Press: e119--e119.↩
[R3] Stanke, Mario, Mark Diekhans, Robert Baertsch, and David Haussler. 2008. “Using Native and Syntenically Mapped cDNA Alignments to Improve de Novo Gene Finding.” Bioinformatics 24 (5). Oxford University Press: 637--44.↩
[R4] Stanke, Mario, Oliver Schöffmann, Burkhard Morgenstern, and Stephan Waack. 2006. “Gene Prediction in Eukaryotes with a Generalized Hidden Markov Model That Uses Hints from External Sources.” BMC Bioinformatics 7 (1). BioMed Central: 62.↩
[R5] Barnett, Derek W, Erik K Garrison, Aaron R Quinlan, Michael P Strömberg, and Gabor T Marth. 2011. “BamTools: A C++ Api and Toolkit for Analyzing and Managing Bam Files.” Bioinformatics 27 (12). Oxford University Press: 1691--2.↩
[R6] Li, Heng, Handsaker, Bob, Alec Wysoker, Tim Fennell, Jue Ruan, Nils Homer, Gabor Marth, Goncalo Abecasis, and Richard Durbin. 2009. “The Sequence Alignment/Map Format and Samtools.” Bioinformatics 25 (16). Oxford University Press: 2078--9.↩
[R7] Gremme, G. 2013. “Computational Gene Structure Prediction.” PhD thesis, Universität Hamburg.↩
[R8] Gotoh, Osamu. 2008a. “A Space-Efficient and Accurate Method for Mapping and Aligning cDNA Sequences onto Genomic Sequence.” Nucleic Acids Research 36 (8). Oxford University Press: 2630--8.↩
[R9] Iwata, Hiroaki, and Osamu Gotoh. 2012. “Benchmarking Spliced Alignment Programs Including Spaln2, an Extended Version of Spaln That Incorporates Additional Species-Specific Features.” Nucleic Acids Research 40 (20). Oxford University Press: e161--e161.↩
[R10] Osamu Gotoh. 2008b。 “Direct Mapping and Alignment of Protein Sequences onto Genomic Sequence.” Bioinformatics 24 (21). Oxford University Press: 2438--44.↩
[R11] Slater, Guy St C, and Ewan Birney. 2005. “Automated Generation of Heuristics for Biological Sequence Comparison.” BMC Bioinformatics 6(1). BioMed Central: 31.↩
[R12] Altschul, SF, W. Gish, W. Miller, EW Myers, and DJ Lipman. 1990. “Basic Local Alignment Search Tool.” Journal of Molecular Biology 215:403--10.↩
[R13] Camacho, Christiam, et al. 2009. “BLAST+: architecture and applications.“ BMC Bioinformatics 1(1): 421.↩
[R14] Lomsadze, A., V. Ter-Hovhannisyan, YO Chernoff, and M. Borodovsky. 2005. “Gene identification in novel eukaryotic genomes by self-training algorithm.” Nucleic Acids Research 33 (20): 6494--6506. doi:10.1093/nar/gki937.↩
[R15] Ter-Hovhannisyan, Vardges, Alexandre Lomsadze, Yury O Chernoff, and Mark Borodovsky. 2008. “Gene Prediction in Novel Fungal Genomes Using an Ab Initio Algorithm with Unsupervised Training.”基因组研究。 Cold Spring Harbor Lab, gr--081612.↩
[R16] Hoff, KJ 2019. MakeHub: Fully automated generation of UCSC Genome Browser Assembly Hubs. Genomics, Proteomics and Bioinformatics , in press, preprint on bioarXive, doi: https://doi.org/10.1101/550145.↩
[R17] Bruna, T., Lomsadze, A., & Borodovsky, M. 2020. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics and Bioinformatics, 2(2), lqaa026. doi: https://doi.org/10.1093/nargab/lqaa026.↩
[R18] Kriventseva, EV, Kuznetsov, D., Tegenfeldt, F., Manni, M., Dias, R., Simão, FA, and Zdobnov, EM 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Research, 47(D1), D807-D811.↩
[R19] Keilwagen, J., Hartung, F., Grau, J. (2019) GeMoMa: Homology-based gene prediction utilizing intron position conservation and RNA-seq data.方法摩尔生物。 1962:161-177, doi: 10.1007/978-1-4939-9173-0_9.↩
[R20] Keilwagen, J., Wenk, M., Erickson, JL, Schattat, MH, Grau, J., Hartung F. (2016) Using intron position conservation for homology-based gene prediction. Nucleic Acids Research, 44(9):e89.↩
[R21] Keilwagen, J., Hartung, F., Paulini, M., Twardziok, SO, Grau, J. (2018) Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics, 19(1):189.↩
[R22] SRA Toolkit Development Team (2020). SRA Toolkit. https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software.[↩](#a22)
[R23] Kim, D., Paggi, JM, Park, C., Bennett, C., & Salzberg, SL (2019).基于图的基因组对齐和基因分型与HISAT2和HISAT基因型。 Nature biotechnology, 37(8):907-915.↩
[R24] Quinlan, AR (2014). BEDTools: the Swiss‐army tool for genome feature analysis. Current protocols in bioinformatics, 47(1):11-12.↩
[R25] Kovaka, S., Zimin, AV, Pertea, GM, Razaghi, R., Salzberg, SL, & Pertea, M. (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome biology, 20(1):1-13.↩
[R26] Pertea, G., & Pertea, M. (2020). GFF utilities: GffRead and GffCompare. F1000Research, 9.↩
[R27] Huang, N., & Li, H. (2023). compleasm: a faster and more accurate reimplementation of BUSCO. Bioinformatics, 39(10), btad595.↩
[R28] Bruna, T., Gabriel, L. & Hoff, KJ (2024). Navigating Eukaryotic Genome Annotation Pipelines: A Route Map to BRAKER, Galba, and TSEBRA. arXiv, https://doi.org/10.48550/arXiv.2403.19416 .↩