Meme Suite工具家族的R接口,该界面为DNA,RNA和蛋白质序列提供了多个实用程序。模因通过检测模因套件的本地安装,运行命令,然后将结果直接导入R。
Memes目前可在Bioconductor devel分支上找到:
if ( ! requireNamespace( " BiocManager " , quietly = TRUE ))
install.packages( " BiocManager " )
# The following initializes usage of Bioc devel
BiocManager :: install( version = ' devel ' )
BiocManager :: install( " memes " )您可以从GitHub安装Memes的开发版本:
if ( ! requireNamespace( " remotes " , quietly = TRUE ))
install.packages( " remotes " )
remotes :: install_github( " snystrom/memes " )
# To temporarily bypass the R version 4.1 requirement, you can pull from the following branch:
remotes :: install_github( " snystrom/memes " , ref = " no-r-4 " ) # Get development version from dockerhub
docker pull snystrom/memes_docker:devel
# the -v flag is used to mount an analysis directory,
# it can be excluded for demo purposes
docker run -e PASSWORD= < password > -p 8787:8787 -v < path > / < to > / < project > :/mnt/ < project > snystrom/memes_docker:devel模因依靠模因套件的本地安装。有关Meme Suite的安装说明,请参见Meme Suite安装指南。
模因需要知道您本地机器上meme/bin/目录的位置。您可以通过4种方式告诉MEME MEME SUITE安装的位置。如果模因是有效的路径,则始终更喜欢更具体的定义。在这里,它们的排名是最不具体的:
meme_path参数options(meme_bin = "/path/to/meme/bin/")设置路径MEME_BIN=/path/to/meme/bin/在您的.Renviron文件中~/meme/bin/如果模因未能在指定位置检测您的安装,则将落回下一个选项。
要验证模因可以检测您的模因安装,请使用check_meme_install()使用上面的搜索herirarchy查找有效的模因安装。它将报告是否缺少任何工具,并打印出看到的模因的路径。这对于解决安装问题的故障排除可能很有用。
library( memes )
# Verify that memes detects your meme install
# (returns all green checks if so)
check_meme_install()
# > checking main install
# > ✓ /opt/meme/bin
# > checking util installs
# > ✓ /opt/meme/bin/dreme
# > ✓ /opt/meme/bin/ame
# > ✓ /opt/meme/bin/fimo
# > ✓ /opt/meme/bin/tomtom
# > ✓ /opt/meme/bin/meme
# > x /opt/meme/bin/streme # You can manually input a path to meme_path
# If no meme/bin is detected, will return a red X
check_meme_install( meme_path = ' bad/path ' )
# > checking main install
# > x bad/path | 功能名称 | 使用 | 序列输入 | 主题输入 | 输出 |
|---|---|---|---|---|
runStreme() | 图案发现(简短主题) | 是的 | 不 | universalmotif_df |
runDreme() | 图案发现(简短主题) | 是的 | 不 | universalmotif_df |
runAme() | 图案富集 | 是的 | 是的 | data.frame(可选: sequences列) |
runFimo() | 主题扫描 | 是的 | 是的 | 主题位置的granges |
runTomTom() | 主题比较 | 不 | 是的 | universalmotif_df w/ best_match_motif和tomtom列* |
runMeme() | 图案发现(长图案) | 是的 | 不 | universalmotif_df |
*注意:如果使用universalmotif_df运行runTomTom()则结果将与universalmotif_df结果作为额外的列连接。这可以轻松地比较De-Novo发现的主题与它们的比赛。
序列输入可以是:
Biostrings::XStringSet (可以使用get_sequence()辅助功能从granges生成)Biostrings::XStringSet对象(由get_sequence()生成)主题输入可以是:
universalmotif对象或universalmotif对象列表runDreme()结果对象(这允许runDreme()的结果直接传递到runTomTom() )list() (例如list("path/to/database.meme", "dreme_results" = dreme_res) )输出类型:
runDreme() , runStreme() , runMeme()和runTomTom()返回带有特殊列的data.frames universalmotif_df对象。 motif列包含一个universalmotif对象,每行1个条目。其余列描述了每个返回基序的属性。以下列名是特殊的,因为在运行update_motifs()和to_list()以更改存储在motif列中的图案的属性时,使用它们的值。在调用update_motifs()或to_list()时,请谨慎更改这些值,因为这些更改将传播到motif列。
模因围绕UniversalMotif软件包构建,该软件包为R. universalmotif_df对象中的操纵图案提供了一个框架,可以分别使用to_df()和to_list()和to_list()函数在data.frame和universalmotif列表格式之间进行互连。这允许使用所有其他Bioconductor图案软件包使用memes结果,因为universalmotif对象可以使用convert_motifs()转换为任何其他主题类型。
runTomTom()返回一个特殊列: tomtom ,这是每个输入基线的所有匹配数据的data.frame 。可以使用tidyr::unnest(tomtom_results, "tomtom")扩展这一点,并用nest_tomtom()重新定位。由runTomTom()返回的best_match_前缀列指示图案的值,这是与输入图案的最佳匹配。
suppressPackageStartupMessages(library( magrittr ))
suppressPackageStartupMessages(library( GenomicRanges ))
# Example transcription factor peaks as GRanges
data( " example_peaks " , package = " memes " )
# Genome object
dm.genome <- BSgenome.Dmelanogaster.UCSC.dm6 :: BSgenome.Dmelanogaster.UCSC.dm6 get_sequence函数将GRanges或GRangesList作为输入,并分别返回序列作为BioStrings::XStringSet或XStringSet对象列表。 get_sequence将以每个序列来自的基因组坐标来命名每个FASTA条目。
# Generate sequences from 200bp about the center of my peaks of interest
sequences <- example_peaks % > %
resize( 200 , " center " ) % > %
get_sequence( dm.genome ) runDreme()接受Xstringset或FastA文件作为输入的路径。您可以将其他序列或洗牌输入序列作为控制数据集。
# runDreme accepts all arguments that the commandline version of dreme accepts
# here I set e = 50 to detect motifs in the limited example peak list
# In a real analysis, e should typically be < 1
dreme_results <- runDreme( sequences , control = " shuffle " , e = 50 )模因围绕环球软件包构建。结果以universalmotif_df格式返回,该格式是R data.Frame,可以使用to_list()在data.frame和universalmotif格式之间无缝互连以将其转换为universalmotif列表格式,而to_df()则可以转换回data.frame格式。使用to_list()允许使用所有universalmotif函数使用memes结果:
library( universalmotif )
dreme_results % > %
to_list() % > %
view_motifs()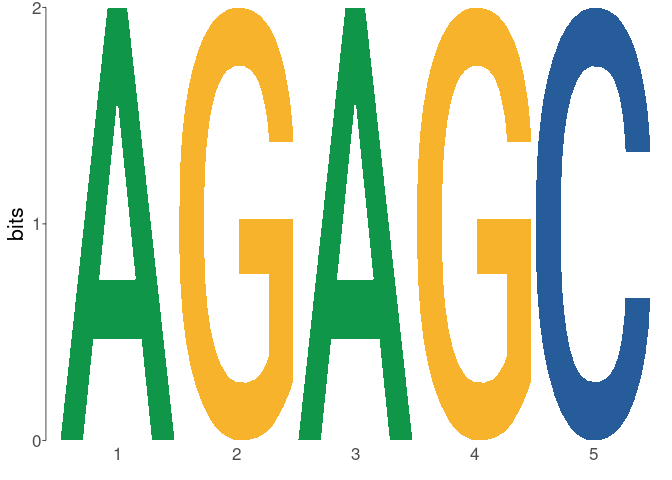
可以使用runTomTom()将发现的主题与已知的TF主题匹配,后者可以作为输入的输入路径,通往.meme格式化文件, universalmotif列表或runDreme()的结果。
TomTom使用已知基础的数据库,可以将其传递到database参数作为.meme格式文件或universalmotif对象的路径。
可选地,您可以将Environment变量MEME_DB设置为.Renviron中的renviron中的文件,或将options中的meme_db值设置为有效的.meme格式文件和memes将使用该文件作为数据库。模因始终更喜欢用户输入而不是全局变量设置。
options( meme_db = system.file( " extdata/flyFactorSurvey_cleaned.meme " , package = " memes " ))
m <- create_motif( " CMATTACN " , altname = " testMotif " )
tomtom_results <- runTomTom( m ) tomtom_results
# > motif name altname consensus alphabet strand icscore type
# > 1 <mot:motif> motif testMotif CMATTACN DNA +- 13 PPM
# > bkg best_match_name best_match_altname
# > 1 0.25, 0.25, 0.25, 0.25 prd_FlyReg prd
# > best_db_name best_match_offset best_match_pval best_match_eval
# > 1 flyFactorSurvey_cleaned 0 9.36e-05 0.052
# > best_match_qval best_match_strand
# > 1 0.0353 +
# > best_match_motif
# > 1 <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>
#> tomtom
#> 1 prd_FlyReg, tup_SOLEXA_10, CG13424_Cell, CG11085_Cell, BH2_Cell, CG13424_SOLEXA_2, Tup_Cell, Tup_SOLEXA, Bsh_Cell, Exex_SOLEXA, Odsh_SOLEXA, Unc4_Cell, Ubx_FlyReg, Unc4_SOLEXA, E5_Cell, inv_SOLEXA_5, BH2_SOLEXA, Zen_SOLEXA, CG33980_SOLEXA_2_10, BH1_SOLEXA, CG33980_SOLEXA_2_0, Hgtx_Cell, NK7.1_Cell, Slou_Cell, CG13424_SOLEXA, Zen2_Cell, AbdA_SOLEXA, Antp_SOLEXA, Btn_Cell, Dfd_SOLEXA, Eve_SOLEXA, Ftz_Cell, Hmx_SOLEXA, Hmx_Cell, CG34031_Cell, zen2_SOLEXA_2, En_Cell, Pb_SOLEXA, Slou_SOLEXA, Unpg_Cell, inv_SOLEXA_2, ovo_FlyReg, lim_SOLEXA_2, C15_SOLEXA, Ems_Cell, Btn_SOLEXA, Unpg_SOLEXA, Pb_Cell, Bsh_SOLEXA, Scr_SOLEXA, Zen2_SOLEXA, CG34031_SOLEXA, Eve_Cell, Pph13_Cell, BH1_Cell, CG11085_SOLEXA, CG32532_Cell, en_FlyReg, Dll_SOLEXA, Dfd_Cell, Dr_SOLEXA, Ap_Cell, Ro_Cell, CG4136_SOLEXA, CG33980_SOLEXA, Hbn_SOLEXA, Lbl_Cell, Otp_Cell, Rx_Cell, CG32532_SOLEXA, NK7.1_SOLEXA, Dr_Cell, Odsh_Cell, Al_SOLEXA, Antp_Cell, Hgtx_SOLEXA, Ftz_SOLEXA, Lab_SOLEXA, Dfd_FlyReg, Ap_SOLEXA, Awh_SOLEXA, CG11294_SOLEXA, CG4136_Cell, E5_SOLEXA, Ro_SOLEXA, PhdP_SOLEXA, CG12361_SOLEXA_2, Ind_Cell, Scr_Cell, CG9876_Cell, CG18599_Cell, CG9876_SOLEXA, Otp_SOLEXA, Lbl_SOLEXA, Ubx_Cell, Ubx_SOLEXA, en_SOLEXA_2, Pph13_SOLEXA, Rx_SOLEXA, CG15696_SOLEXA, CG18599_SOLEXA, Ems_SOLEXA, Repo_Cell, Dll_Cell, C15_Cell, CG12361_SOLEXA, Abd-A_FlyReg, Repo_SOLEXA, Zen_Cell, Inv_Cell, En_SOLEXA, Lim3_Cell, Lim1_SOLEXA, CG15696_Cell, Crc_CG6272_SANGER_5, Lab_Cell, CG32105_SOLEXA, Bap_SOLEXA, CG9437_SANGER_5, AbdA_Cell, pho_FlyReg, CG33980_Cell, Cad_SOLEXA, CG4328_SOLEXA, CG4328_Cell, Gsc_Cell, vri_SANGER_5, AbdB_SOLEXA, Xrp1_CG6272_SANGER_5, Al_Cell, Exex_Cell, br-Z4_FlyReg, CG11294_Cell, Aef1_FlyReg, CG7745_SANGER_5, PhdP_Cell, Awh_Cell, prd, tup, lms, CG11085, B-H2, lms, tup, tup, bsh, exex, OdsH, unc-4, Ubx, unc-4, E5, inv, B-H2, zen, CG33980, B-H1, CG33980, HGTX, NK7.1, slou, lms, zen2, abd-A, Antp, btn, Dfd, eve, ftz, Hmx, Hmx, CG34031, zen2, en, pb, slou, unpg, inv, ovo, Lim1, C15, ems, btn, unpg, pb, bsh, Scr, zen2, CG34031, eve, Pph13, B-H1, CG11085, CG32532, en, Dll, Dfd, Dr, ap, ro, CG4136, CG33980, hbn, lbl, otp, Rx, CG32532, NK7.1, Dr, OdsH, al, Antp, HGTX, ftz, lab, Dfd, ap, Awh, CG11294, CG4136, E5, ro, PHDP, CG12361, ind, Scr, CG9876, CG18599, CG9876, otp, lbl, Ubx, Ubx, en, Pph13, Rx, CG15696, CG18599, ems, repo, Dll, C15, CG12361, abd-A, repo, zen, inv, en, Lim3, Lim1, CG15696, crc, lab, CG32105, bap, CG9437, abd-A, pho, CG33980, cad, CG4328, CG4328, Gsc, vri, Abd-B, Xrp1, al, exex, br, CG11294, Aef1, CG7745, PHDP, Awh, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, <S4 class 'universalmotif' [package "universalmotif"] with 20 slots>, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, flyFactorSurvey_cleaned, 0, 1, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, -1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, -2, 0, 0, 0, -3, 0, 4, 1, 0, 0, 0, 0, -2, 0, -2, 1, 0, -2, 1, 0, 2, -1, 0, 9.36e-05, 0.000129, 0.00013, 0.000194, 0.000246, 0.000247, 0.000319, 0.000364, 0.000427, 0.000435, 0.000473, 0.000473, 0.000556, 0.000643, 0.000694, 0.000728, 0.000748, 0.000748, 0.000785, 0.000828, 0.000849, 0.000868, 0.000868, 0.000868, 0.00102, 0.00113, 0.00116, 0.00116, 0.00116, 0.00116, 0.00116, 0.00116, 0.00118, 0.00126, 0.00134, 0.00141, 0.00144, 0.00145, 0.00145, 0.00154, 0.00161, 0.00165, 0.00173, 0.00177, 0.00177, 0.00179, 0.00179, 0.0019, 0.00191, 0.00191, 0.00191, 0.00203, 0.00203, 0.00203, 0.00212, 0.00214, 0.00219, 0.00232, 0.00242, 0.00247, 0.0026, 0.00264, 0.00264, 0.00268, 0.00282, 0.00282, 0.00282, 0.00282, 0.00282, 0.00286, 0.00286, 0.00296, 0.00305, 0.00316, 0.0032, 0.0032, 0.00326, 0.00326, 0.00341, 0.00348, 0.00348, 0.00348, 0.00348, 0.00348, 0.00348, 0.0035, 0.00359, 0.00359, 0.00359, 0.00364, 0.00372, 0.00372, 0.00372, 0.00387, 0.00387, 0.00412, 0.00415, 0.00424, 0.00424, 0.00439, 0.00452, 0.00452, 0.00467, 0.00483, 0.00497, 0.00497, 0.00528, 0.00549, 0.0059, 0.00597, 0.00624, 0.00635, 0.00709, 0.00761, 0.0079, 0.00856, 0.00859, 0.00912, 0.00935, 0.0097, 0.00974, 0.0101, 0.0109, 0.0116, 0.0123, 0.0123, 0.0127, 0.0138, 0.0138, 0.014, 0.014, 0.0147, 0.0148, 0.0155, 0.0165, 0.0166, 0.0177, 0.052, 0.0718, 0.0725, 0.108, 0.137, 0.137, 0.177, 0.202, 0.238, 0.242, 0.263, 0.263, 0.309, 0.357, 0.386, 0.405, 0.416, 0.416, 0.436, 0.46, 0.472, 0.483, 0.483, 0.483, 0.566, 0.629, 0.646, 0.646, 0.646, 0.646, 0.646, 0.646, 0.654, 0.702, 0.745, 0.785, 0.8, 0.808, 0.808, 0.858, 0.897, 0.92, 0.961, 0.985, 0.985, 0.993, 0.993, 1.05, 1.06, 1.06, 1.06, 1.13, 1.13, 1.13, 1.18, 1.19, 1.22, 1.29, 1.34, 1.38, 1.45, 1.47, 1.47, 1.49, 1.57, 1.57, 1.57, 1.57, 1.57, 1.59, 1.59, 1.65, 1.7, 1.76, 1.78, 1.78, 1.81, 1.81, 1.9, 1.94, 1.94, 1.94, 1.94, 1.94, 1.94, 1.95, 2, 2, 2, 2.02, 2.07, 2.07, 2.07, 2.15, 2.15, 2.29, 2.31, 2.36, 2.36, 2.44, 2.52, 2.52, 2.6, 2.68, 2.76, 2.76, 2.94, 3.05, 3.28, 3.32, 3.47, 3.53, 3.94, 4.23, 4.39, 4.76, 4.77, 5.07, 5.2, 5.39, 5.41, 5.61, 6.06, 6.42, 6.81, 6.81, 7.03, 7.66, 7.65, 7.78, 7.78, 8.15, 8.26, 8.59, 9.18, 9.2, 9.85, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0353, 0.0368, 0.0369, 0.0369, 0.0369, 0.0369, 0.0369, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0372, 0.0379, 0.0379, 0.0381, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0396, 0.0404, 0.0404, 0.0424, 0.0424, 0.0424, 0.0424, 0.0435, 0.0439, 0.0439, 0.0449, 0.046, 0.0464, 0.0464, 0.0489, 0.0504, 0.0536, 0.0538, 0.0557, 0.0562, 0.0622, 0.0662, 0.0681, 0.0727, 0.0727, 0.0765, 0.0778, 0.0797, 0.0797, 0.0819, 0.0877, 0.0923, 0.0964, 0.0964, 0.0987, 0.106, 0.106, 0.106, 0.106, 0.11, 0.111, 0.114, 0.121, 0.121, 0.128, +, -, -, -, -, -, -, -, -, -, -, -, +, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, +, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, +, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, -, +, -, -, -, -, -, -, -, +, -, -, -, +, -, -, -, -, -, -, -, +, -, +, -, -, -, +, +, +, -, -
# >
# > [Hidden empty columns: family, organism, nsites, bkgsites, pval, qval,
# > eval.]runTomTom()将其结果作为列将其结果添加到runDreme()结果data.frame。
full_results <- dreme_results % > %
runTomTom()AME用于测试目标序列中已知基序的富集。 runAme()将使用.Renviron或options(meme_db = "path/to/database.meme")中的MEME_DB条目作为主题数据库。或者,它将接受类似于runTomTom()类似的所有有效输入。
# here I set the evalue_report_threshold = 30 to detect motifs in the limited example sequences
# In a real analysis, evalue_report_threshold should be carefully selected
ame_results <- runAme( sequences , control = " shuffle " , evalue_report_threshold = 30 )
ame_results
# > # A tibble: 2 x 17
# > rank motif_db motif_id motif_alt_id consensus pvalue adj.pvalue evalue tests
# > <int> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <int>
# > 1 1 /usr/lo… Eip93F_… Eip93F ACWSCCRA… 5.14e-4 0.0339 18.8 67
# > 2 2 /usr/lo… Cf2-PB_… Cf2 CSSHNKDT… 1.57e-3 0.0415 23.1 27
# > # … with 8 more variables: fasta_max <dbl>, pos <int>, neg <int>,
# > # pwm_min <dbl>, tp <int>, tp_percent <dbl>, fp <int>, fp_percent <dbl> view_tomtom_hits允许将输入图案与TomTom的最高点击进行比较。对这些匹配的手动检查很重要,因为有时最重要的比赛并不总是正确的分配。更改top_n可以使您以排名的降序显示其他匹配项。
full_results % > %
view_tomtom_hits( top_n = 1 )
# > $m01_AGAGC 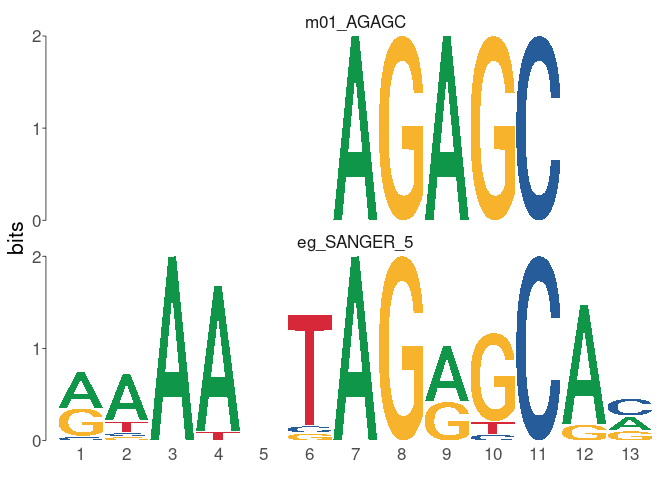
将runAme()的结果视为热图可能很有用。 plot_ame_heatmap()可以创建复杂的可视化,以分析不同区域类型之间的富集(有关详细信息,请参见小插图)。这是一个简单的示例热图。
ame_results % > %
plot_ame_heatmap()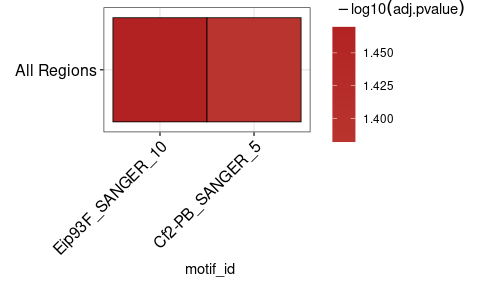
FIMO工具用于识别与已知主题的匹配。 runFimo将作为包含基匹配基因组坐标的GRanges对象返回这些命中。
# Query MotifDb for a motif
e93_motif <- MotifDb :: query( MotifDb :: MotifDb , " Eip93F " ) % > %
universalmotif :: convert_motifs()
# > See system.file("LICENSE", package="MotifDb") for use restrictions.
# Scan for the E93 motif within given sequences
fimo_results <- runFimo( sequences , e93_motif , thresh = 1e-3 )
# Visualize the sequences matching the E93 motif
plot_sequence_heatmap( fimo_results $ matched_sequence ) 
模因还支持使用R外部的Meme Suite生成的导入结果(例如,在meme-suite.org上运行作业或在命令行上运行)。这使得使用先前存在的模因套件结果具有下游模因函数。
| 模因工具 | 功能名称 | 文件类型 |
|---|---|---|
| streme | importStremeXML() | streme.xml |
| Dreme | importDremeXML() | dreme.xml |
| Tomtom | importTomTomXML() | tomtom.xml |
| AME | importAme() | ame.tsv* |
| fimo | importFimo() | fimo.tsv |
| 模因 | importMeme() | meme.txt |
* importAME()还可以在AME使用method = "fisher"时使用“ sequences.tsv”输出,这是可选的。
该模因套件当前不支持Windows,尽管可以在Cygwin或Windows Linux Subsytem(WSL)下安装。请注意,如果将模因安装在Cygwin或WSL上,则还必须在Cygwin或WSL内运行R才能使用模因。
另一种解决方案是使用Docker安装模因套件来运行虚拟环境。我们提供一个模因码头容器
与模因套房,R Studio和所有memes依赖关系预装在一起。
Memes是来自Meme Suite的一些选择工具的包装,该工具是由另一组开发的。除了引用模因之外,请引用与您使用的工具相对应的模因套件工具。
如果您在分析中使用runDreme() ,请引用:
蒂莫西·贝利(Timothy L.
如果您在分析中使用runTomTom() ,请引用:
Shobhit Gupta,Ja Stamatoyannopolous,Timothy Bailey和William Stafford Noble,“量化图案之间的相似性”,Genome Biology,8(2):R24,2007。全文。
如果您在分析中使用runAme() ,请引用:
Robert McLeay和Timothy L. Bailey,“主题富集分析:统一框架和方法评估”,BMC BioInformatics,11:165,2010,doi:10.1186/1471-2105-11-165。全文
如果您在分析中使用runFimo() ,请引用:
查尔斯·E·格兰特(Charles E.
Meme Suite是免费的,可用于非营利性使用,但营利性用户应购买许可证。有关详细信息,请参见Meme Suite版权页面。