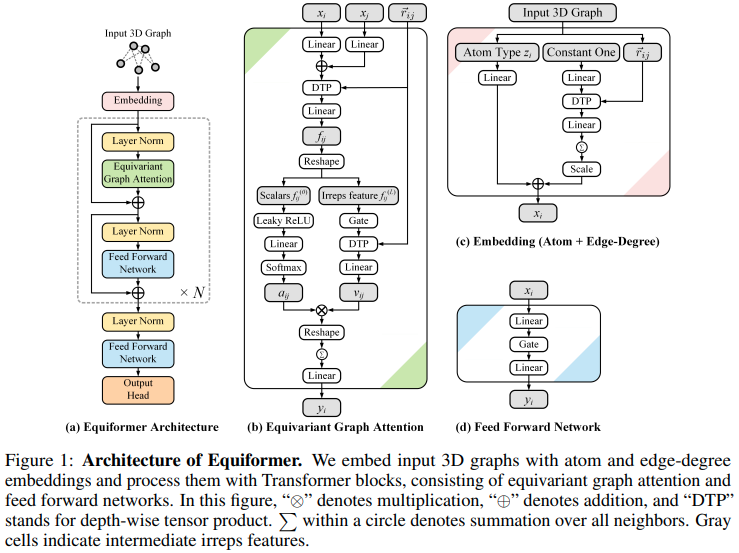
實現 Equiformer、SE3/E3 等變注意力網絡,達到新的 SOTA,並被 EquiFold(Prescient Design)採用用於蛋白質折疊
這個設計似乎是建立在 SE3 Transformers 的基礎上的,用 MLP Attention 和 GATv2 的非線性訊息傳遞取代了點積注意力。它還進行深度張量乘積以提高效率。如果您認為我錯了,請隨時給我發電子郵件。
更新:有一項新的進展,讓 SE3 等變網路的度數擴展變得更好!本文首先指出,沿著 z 軸(或以其他慣例為 y 軸)對齊表示,球諧函數會變得稀疏。這從方程式中刪除了 m f維度。 Passaro 等人的後續論文。注意到 Clebsch Gordan 矩陣也變得稀疏,導致mi和 l f被刪除。他們還認為,在將重複次數與一個軸對齊後,問題已從 SO(3) 減少到 SO(2)。 Equiformer v2(官方儲存庫)在類似 Transformer 的框架中利用這一點來達到新的 SOTA。
肯定會投入更多的工作/探索。現在,我已經將前兩篇論文中的技巧融入 Equiformer v1 中,除了完全轉換為 SO(2) 之外。
更新 2:似乎有一個新的 SOTA,在更高程度的代表之間沒有任何交互(換句話說,所有張量積/clebsch gordan 數學都消失了)。 GotenNet,似乎是 HEGNN 的變形版
$ pip install equiformer-pytorch import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 24 ,
dim = ( 4 , 4 , 2 ), # dimensions per type, ascending, length must match number of degrees (num_degrees)
dim_head = ( 4 , 4 , 4 ), # dimension per attention head
heads = ( 2 , 2 , 2 ), # number of attention heads
num_linear_attn_heads = 0 , # number of global linear attention heads, can see all the neighbors
num_degrees = 3 , # number of degrees
depth = 4 , # depth of equivariant transformer
attend_self = True , # attending to self or not
reduce_dim_out = True , # whether to reduce out to dimension of 1, say for predicting new coordinates for type 1 features
l2_dist_attention = False # set to False to try out MLP attention
). cuda ()
feats = torch . randint ( 0 , 24 , ( 1 , 128 )). cuda ()
coors = torch . randn ( 1 , 128 , 3 ). cuda ()
mask = torch . ones ( 1 , 128 ). bool (). cuda ()
out = model ( feats , coors , mask ) # (1, 128)
out . type0 # invariant type 0 - (1, 128)
out . type1 # equivariant type 1 - (1, 128, 3)該儲存庫還包括一種使用可逆網路將記憶體使用與深度解耦的方法。換句話說,如果增加深度,則使用等量變換器區塊(注意力和前饋)時,記憶體成本將保持不變。
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 24 ,
dim = ( 4 , 4 , 2 ),
dim_head = ( 4 , 4 , 4 ),
heads = ( 2 , 2 , 2 ),
num_degrees = 3 ,
depth = 48 , # depth of 48 - just to show that it runs - in reality, seems to be quite unstable at higher depths, so architecture stil needs more work
reversible = True , # just set this to True to use https://arxiv.org/abs/1707.04585
). cuda ()
feats = torch . randint ( 0 , 24 , ( 1 , 128 )). cuda ()
coors = torch . randn ( 1 , 128 , 3 ). cuda ()
mask = torch . ones ( 1 , 128 ). bool (). cuda ()
out = model ( feats , coors , mask )
out . type0 . sum (). backward ()有邊緣,例如。原子鍵
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
num_tokens = 28 ,
dim = 64 ,
num_edge_tokens = 4 , # number of edge type, say 4 bond types
edge_dim = 16 , # dimension of edge embedding
depth = 2 ,
input_degrees = 1 ,
num_degrees = 3 ,
reduce_dim_out = True
)
atoms = torch . randint ( 0 , 28 , ( 2 , 32 ))
bonds = torch . randint ( 0 , 4 , ( 2 , 32 , 32 ))
coors = torch . randn ( 2 , 32 , 3 )
mask = torch . ones ( 2 , 32 ). bool ()
out = model ( atoms , coors , mask , edges = bonds )
out . type0 # (2, 32)
out . type1 # (2, 32, 3)與鄰接矩陣
import torch
from equiformer_pytorch import Equiformer
model = Equiformer (
dim = 32 ,
heads = 8 ,
depth = 1 ,
dim_head = 64 ,
num_degrees = 2 ,
valid_radius = 10 ,
reduce_dim_out = True ,
attend_sparse_neighbors = True , # this must be set to true, in which case it will assert that you pass in the adjacency matrix
num_neighbors = 0 , # if you set this to 0, it will only consider the connected neighbors as defined by the adjacency matrix. but if you set a value greater than 0, it will continue to fetch the closest points up to this many, excluding the ones already specified by the adjacency matrix
num_adj_degrees_embed = 2 , # this will derive the second degree connections and embed it correctly
max_sparse_neighbors = 8 # you can cap the number of neighbors, sampled from within your sparse set of neighbors as defined by the adjacency matrix, if specified
)
feats = torch . randn ( 1 , 128 , 32 )
coors = torch . randn ( 1 , 128 , 3 )
mask = torch . ones ( 1 , 128 ). bool ()
# placeholder adjacency matrix
# naively assuming the sequence is one long chain (128, 128)
i = torch . arange ( 128 )
adj_mat = ( i [:, None ] <= ( i [ None , :] + 1 )) & ( i [:, None ] >= ( i [ None , :] - 1 ))
out = model ( feats , coors , mask , adj_mat = adj_mat )
out . type0 # (1, 128)
out . type1 # (1, 128, 3) 等方差測試等
$ python setup.py test 首先安裝sidechainnet
$ pip install sidechainnet然後運行蛋白質主幹去雜訊任務
$ python denoise.py將 xi 和 xj 單獨的項目和求和邏輯移至 Conv 類別中
將自交互鍵/值生成移至Conv,修復自交互中的無池化問題
採用簡單的方法來分割 DTP 輸入程度的貢獻
對於更高類型的點積注意力,請嘗試歐幾里德距離
考慮僅針對 type0 的所有鄰居注意力層,使用線性注意力
整合球形通道論文中的新發現,隨後是 so(3) -> so(2) 論文,這減少了 O(L^6) -> O(L^3) 的計算!
@article { Liao2022EquiformerEG ,
title = { Equiformer: Equivariant Graph Attention Transformer for 3D Atomistic Graphs } ,
author = { Yi Liao and Tess E. Smidt } ,
journal = { ArXiv } ,
year = { 2022 } ,
volume = { abs/2206.11990 }
} @article { Lee2022.10.07.511322 ,
author = { Lee, Jae Hyeon and Yadollahpour, Payman and Watkins, Andrew and Frey, Nathan C. and Leaver-Fay, Andrew and Ra, Stephen and Cho, Kyunghyun and Gligorijevic, Vladimir and Regev, Aviv and Bonneau, Richard } ,
title = { EquiFold: Protein Structure Prediction with a Novel Coarse-Grained Structure Representation } ,
elocation-id = { 2022.10.07.511322 } ,
year = { 2022 } ,
doi = { 10.1101/2022.10.07.511322 } ,
publisher = { Cold Spring Harbor Laboratory } ,
URL = { https://www.biorxiv.org/content/early/2022/10/08/2022.10.07.511322 } ,
eprint = { https://www.biorxiv.org/content/early/2022/10/08/2022.10.07.511322.full.pdf } ,
journal = { bioRxiv }
} @article { Shazeer2019FastTD ,
title = { Fast Transformer Decoding: One Write-Head is All You Need } ,
author = { Noam M. Shazeer } ,
journal = { ArXiv } ,
year = { 2019 } ,
volume = { abs/1911.02150 }
} @misc { ding2021cogview ,
title = { CogView: Mastering Text-to-Image Generation via Transformers } ,
author = { Ming Ding and Zhuoyi Yang and Wenyi Hong and Wendi Zheng and Chang Zhou and Da Yin and Junyang Lin and Xu Zou and Zhou Shao and Hongxia Yang and Jie Tang } ,
year = { 2021 } ,
eprint = { 2105.13290 } ,
archivePrefix = { arXiv } ,
primaryClass = { cs.CV }
} @inproceedings { Kim2020TheLC ,
title = { The Lipschitz Constant of Self-Attention } ,
author = { Hyunjik Kim and George Papamakarios and Andriy Mnih } ,
booktitle = { International Conference on Machine Learning } ,
year = { 2020 }
} @article { Zitnick2022SphericalCF ,
title = { Spherical Channels for Modeling Atomic Interactions } ,
author = { C. Lawrence Zitnick and Abhishek Das and Adeesh Kolluru and Janice Lan and Muhammed Shuaibi and Anuroop Sriram and Zachary W. Ulissi and Brandon C. Wood } ,
journal = { ArXiv } ,
year = { 2022 } ,
volume = { abs/2206.14331 }
} @article { Passaro2023ReducingSC ,
title = { Reducing SO(3) Convolutions to SO(2) for Efficient Equivariant GNNs } ,
author = { Saro Passaro and C. Lawrence Zitnick } ,
journal = { ArXiv } ,
year = { 2023 } ,
volume = { abs/2302.03655 }
} @inproceedings { Gomez2017TheRR ,
title = { The Reversible Residual Network: Backpropagation Without Storing Activations } ,
author = { Aidan N. Gomez and Mengye Ren and Raquel Urtasun and Roger Baker Grosse } ,
booktitle = { NIPS } ,
year = { 2017 }
} @article { Bondarenko2023QuantizableTR ,
title = { Quantizable Transformers: Removing Outliers by Helping Attention Heads Do Nothing } ,
author = { Yelysei Bondarenko and Markus Nagel and Tijmen Blankevoort } ,
journal = { ArXiv } ,
year = { 2023 } ,
volume = { abs/2306.12929 } ,
url = { https://api.semanticscholar.org/CorpusID:259224568 }
} @inproceedings { Arora2023ZoologyMA ,
title = { Zoology: Measuring and Improving Recall in Efficient Language Models } ,
author = { Simran Arora and Sabri Eyuboglu and Aman Timalsina and Isys Johnson and Michael Poli and James Zou and Atri Rudra and Christopher R'e } ,
year = { 2023 } ,
url = { https://api.semanticscholar.org/CorpusID:266149332 }
}